Porcine coronaviruses assessment in manure pits
Information on manure pit testing of pathogens is scarce but may be tool for disease assessment.
May 11, 2021

Porcine coronaviruses have been present in the U.S. swine industry for several years (Stevenson et al., 2013). Transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV) and porcine Deltacoronavirus (PDCoV) continue to be part of the list of differential diagnosis when facing a clinical case of gastrointestinal disease (Marthaler et al., 2014; Turlewicz-Podbielska & Pomorska-Mól, 2021; Xia et al., 2018; Zimmerman., 2019). Unfortunately, both PEDV and PDCoV are responsible for breaks in breeding herds indicating that the virus continues to transmit within the pig population.
Even though the incidence of both viruses in the breeding herd population remains below 10% (MSHMP, 2021), it is not well understood what the situation is in the growing pig herd. Pig production companies continue to invest resources in order to eliminate these viruses from their downstream production flows to aid in the regional control (Drebes, 2020). However, there are still important knowledge gaps in relation to how these viruses continue to infect pig populations.
In an effort to help elucidate the epidemiology of these viruses, we decided to conduct a pilot project with the aim of estimating the prevalence of these coronaviruses in manure pits. Understanding how widespread this virus is and whether it is viable or not can contribute to building stronger biosecurity programs, especially those around manure management activities.
In the fall of 2019, a cross-sectional study was conducted in the Midwest on a total of 300 manure pit samples representing 348 barns in 173 farms belonging to five companies participating in the Morrison Swine Health Monitoring Project (MSHMP). Pit samples were tested at the University of Minnesota Veterinary Diagnostic Laboratory by RT-PCR for the presence of genetic material (RNA) of porcine coronaviruses (e.g., TGEV, PEDV, and PDCoV). Positive samples were then further tested by virus isolation in cell culture to assess whether viruses present in the sample were viable. Samples with the highest viral concentration were then submitted for sequencing.
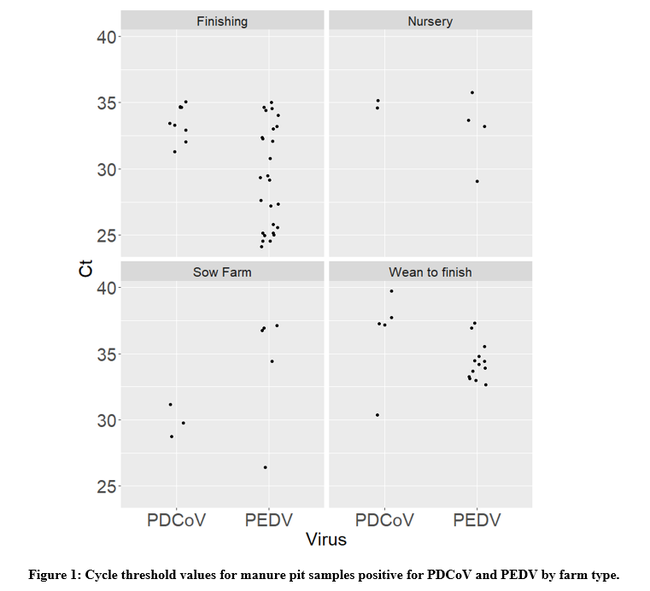
Out of all of the 300 samples tested, PEDV was detected in 48 (16%) farms. When indirectly assessing the amount of virus in the sample through the cycle threshold (ct) value (e.g., lower ct values indicate higher genetic material concentrations), the median ct value was 33. Five samples yielded a ct value ≤25, indicating the samples contained a high concentration of virus RNA. In this dataset, finishing pig farms were the ones that yielded the samples with the highest concentration of PEDV (Figure 1).
In the case of PDCoV, the virus was detected in 19 (6.3%) farms with a median ct value of 34. As with PEDV, a large quantity of growing pig farms was among the positive farms.
Lastly, TGEV was not detected in any of the samples. With regards to virus viability, no viable virus was detected by virus isolation in the tested samples. Samples that were submitted for sequencing did not yield any sequence.
In human medicine, similar work to the one presented here is being conducted with COVID-19 as wastewater-based epidemiology is being used as a surveillance tool (Melvin et al., 2021). Since humans infected with COVID-19 can shed the virus through feces, sampling the wastewater network can provide some estimates that can help understand population shedding of the human coronavirus and thus help inform predictive and transmission models (Michael-Kordatou et al., 2020; Rooney et al., 2021).
In the case of the swine industry, information on manure pit testing of pathogens is scarce. Manure pit sampling protocols do not exist, and more research is needed to understand the significance of a positive result in terms of herd health and also to estimate sensitivity of detection. Since pigs from participating sites were not tested, we cannot make conclusions on whether the virus RNA detected originated from the current or the previous batch of pigs. However, the fact that coronaviruses were detected in manure pits and in some cases at high concentrations may suggest that infected pigs may have been housed recently at this premises. Therefore, results from the current sampling and testing approach may be useful from a monitoring and risk assessment standpoint. Further studies are needed to understand the implications of these findings and further determine the risk of positive manure pits, especially during manure pumping season.
Source: Julian Montoya Lopez, Juan Sanhueza, Carles Vilalta, Cesar Corzo, University of Minnesota, who is solely responsible for the information provided, and wholly owns the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset. The opinions of this writer are not necessarily those of Farm Progress/Informa.
References:
Drebes, D., Robbins, R., Dufresne, L. Investigating PED infections and lessions to improve biosecurity in growing pigs. 2020. Leman Swine Conference.
Marthaler, D., Raymond, L., Jiang, Y., Collins, J., Rossow, K., & Rovira, A. (2014). Rapid Detection, Complete Genome Sequencing, and Phylogenetic Analysis of Porcine Deltacoronavirus. Emerging Infectious Diseases, 20(8), 1347–1350. https://doi.org/10.3201/eid2008.140526
Melvin, R. G., Chaudhry, N., Georgewill, O., Freese, R., & Simmons, G. E. (2021). Predictive power of SARS-CoV-2 wastewater surveillance for diverse populations across a large geographical range [Preprint]. Epidemiology. https://doi.org/10.1101/2021.01.23.21250376
Michael-Kordatou, I., Karaolia, P., & Fatta-Kassinos, D. (2020). Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. Journal of Environmental Chemical Engineering, 8(5), 104306. https://doi.org/10.1016/j.jece.2020.104306
Morrison Swine Health Monitoring Project. 2021.
https://vetmed.umn.edu/centers-programs/swine-program/outreach-leman-mshmp/mshmp/ped-charts
Rooney, C. M., Moura, I. B., & Wilcox, M. H. (2021). Tracking COVID-19 via sewage. Current Opinion in Gastroenterology, 37(1), 4–8. https://doi.org/10.1097/MOG.0000000000000692
Stevenson, G. W., Hoang, H., Schwartz, K. J., Burrough, E. R., Sun, D., Madson, D., Cooper, V. L., Pillatzki, A., Gauger, P., Schmitt, B. J., Koster, L. G., Killian, M. L., & Yoon, K. J. (2013). Emergence of Porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. Journal of Veterinary Diagnostic Investigation, 25(5), 649–654. https://doi.org/10.1177/1040638713501675
Turlewicz-Podbielska, H., & Pomorska-Mól, M. (2021). Porcine Coronaviruses: Overview of the State of the Art. Virologica Sinica, 1–19. https://doi.org/10.1007/s12250-021-00364-0
Xia, L., Yang, Y., Wang, J., Jing, Y., & Yang, Q. (2018). Impact of TGEV infection on the pig small intestine. Virology Journal, 15(1), 102. https://doi.org/10.1186/s12985-018-1012-9
Zimmerman., J. J. (Ed.). (2019). Diseases of swine (11th edition). Wiley-Blackwell.
You May Also Like



