New informative tool for swine veterinarians, producers and stakeholders
SDRS PRRSV BLAST tool allows users to compare their own sequences against the database, identify states, lineage and RFLP of similar sequences.
March 13, 2024

By Srijita Chandra, Guilherme Cezar, Kinath Rupasinghe, Daniel C L Linhares and Giovani Trevisan, Iowa State University
The Swine Disease Reporting System is introducing the SDRS PRRSV BLAST tool, a one-of-a-kind implementation of a Basic Local Alignment Search Tool specific to the PRRSV ORF5 sequences. The tool’s focus is to help answer spatiotemporal questions regarding query sequences and detect any new sequences entering the SDRS database.
The web-based tool allows veterinarians, producers and other users to compare their ORF5 sequences with those in the SDRS database, identify sequences present in the database by level of nucleotide identity, and view where the resulting sequences occurred before, when they occurred, what is the percentage similarity of these sequences with the query sequence as well as the lineage and RFLP of the sequences.
The web-based application (Figure 1) can be found on the SDRS website. The SDRS team can be contacted if additional information or help interpreting the tool's results is required.
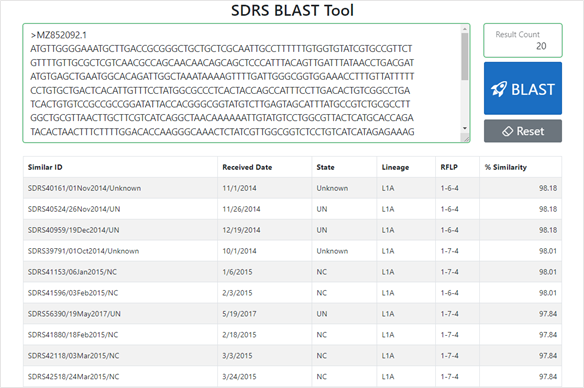
Figure 1: Web interface of the SDRS PRRSV BLAST tool.
Another essential function of the tool is identifying new sequences entering the database. Any sequence with ≤ 6 ambiguities, ≥ 597 base pairs, and nucleotide identity <95% with any pre-existing sequence in the database is flagged as a new sequence, and the SDRS team is notified of these sequences. The tool has identified 133 new sequences since 2010.
Following its initial detection and considering a nucleotide identity of ≥98%, 88.7% (118 of 133) of the sequences had <10 detections, 8.3% (11 of 133) had between 10 and 19, and 3% (4 of 133) had 20 or more occurrences forming a cluster of detections. It can be inferred from the results that 3% of the sequences flagged as new have field relevance. Lineage 5A (15.0%), PRRSV-1 (9.8%) and lineage 1A (8.3%) are the most dominant lineages in these sequences, and the most dominant RFLP patterns are 1-4-4 (12.0%), PRRSV-1 (9.8%) and 1-2-4 (6%).
The tool has an interactive web interface where users can enter their query sequence in a FASTA format and modify the number of results displayed. The result returned has the ID, Received Date, State, Lineage, RFLP and Percentage Similarity columns displayed, which can provide helpful insight into the query sequence's evolution pattern.
Highlights
The SDRS PRRSV BLAST tool is now available to producers, veterinarians, stakeholders, and any other users to compare their own sequences against the SDRS database and identify the states, lineage, and RFLP of similar sequences.
The tool has also been designed to identify new sequences and notify the SDRS team. These sequences will then be reported in the monthly reports.
Funding
This project was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2023-67015-39883 from the USDA’s National Institute of Food and Agriculture.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403-10. doi: 10.1016/S0022-2836(05)80360-2. PMID: 2231712.
Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195-7. doi: 10.1016/0022-2836(81)90087-5. PMID: 7265238.
Zeller MA, Arendsee ZW, Smith GJD, Anderson TK. classLog: Logistic regression for the classification of genetic sequences. bioRxiv, 2022.08.15.503907; doi: https://doi.org/10.1101/2022.08.15.503907.
Yim-Im W, Anderson TK, Paploski IAD, VanderWaal K, Gauger P, Krueger K, Shi M, Main R, Zhang J. Microbiol Spectr. 2023 Nov 7:e0291623. doi: 10.1128/spectrum.02916-23. Online ahead of print. PMID: 37933982.
You May Also Like



