Using gut microbes for respiratory relief
Blueprint: Fecal microbiota transplantation may be preventive strategy for pigs at risk for polymicrobial respiratory disease.
October 29, 2019

By Megan C. Niederwerder, Kansas State University
“Gut microbiome” is the term used to describe the collection of microbes living throughout the length of the gastrointestinal tract. These microbes are vastly diverse and include viruses, bacteria, protozoa, fungi and archaea. Microbes in the gut are estimated to equal or outnumber the cells within the host (Sender et al., 2016).
Importantly, the gut microbiome plays at least three critical roles within the host, including the provision of a protective intestinal barrier which likely prevents pathogen attachment; the digestion and metabolism of nutrients such as complex carbohydrates; and the development, as well as maintenance and regulation, of the immune system.
There are several factors and significant events that affect the initial microbial exposure and colonization of the piglet gut within just the first few weeks of life, including birth route; the vaginal, skin and fecal microbiomes of the sow or gilt; milk and colostrum composition while nursing; the transition to solid feed; housing environment and barn filtration; pathogenic infections; antimicrobial injections and in-feed antimicrobial consumption; and stress at processing or during transportation. Often, these events may play a role in shaping the microbiome for the lifetime of a production pig (Niederwerder, 2017).
The relationship, balance and mechanistic interactions among these microbes in the gut is extremely complex and not well understood in states of host health or disease. However, there is growing evidence supporting the beneficial role that gut microbiome diversity and composition play in the outcome of pathogenic infections.
Alternative strategy
Historically, the impact of the gut microbiome on pathogenic infections has focused on gastroenteric diseases that primarily result in diarrhea, due to the seemingly inherent interactions of beneficial and pathogenic microbes in close proximity within the gut.
For example, pigs characterized as susceptible to enterotoxigenic E. coli had a reduction in intestinal bacterial diversity compared to those pigs considered nonsusceptible (Messori et al., 2013). Similarly, sows that developed diarrhea after natural infection with porcine epidemic diarrhea virus had an overall reduction in observed bacterial species in the gut when compared to sows that remained healthy (Song et al., 2017).
More recently, interest in the gut microbiome and its effects on pathogenic infections has expanded to include pathogens affecting almost all body systems, including those that cause primary pathology in the respiratory tract (Niederwerder, 2017). As polymicrobial respiratory disease is a major cause of morbidity and mortality in growing pigs, the gut microbiome has emerged as an alternative strategy in control and prevention of these infections.
“Gut-lung axis” is the term used to describe how the gut microbiome, likely through products of microbial metabolism and modulation of systemic or mucosal immunity, communicates with the respiratory tract and has the potential to affect outcome following pathogenic respiratory infection or pneumonia.
Several published studies have demonstrated association between microbiome diversity and composition characteristics, with outcome following infection with viral, bacterial and even fungal respiratory pathogens in mice models of human disease (Niederwerder, 2017).
For example, increased gut microbial diversity has been associated with reduced mortality, decreased lung lesions and enhanced alveolar macrophage phagocytic function after respiratory infection with Streptococcus pneumoniae (Schuijt et al., 2016).
Similarly in pigs, increased gut microbial diversity has been associated with reduced clinical signs of Mycoplasma hyopneumoniae infection and decreased lung lesions after respiratory infection with M. hyopneumoniae (Schachtschneider et al., 2013). Furthermore, outcome following infection with respiratory viruses such as influenza virus and respiratory syncytial virus have also been correlated with specific gut microbiome characteristics (Antunes et al., 2019; Rosshart et al., 2017).
PRRS, PCV2 control
Porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 are two of the most important pathogens affecting the swine industry worldwide. Co-infections involving both pathogens are common on a global scale, resulting in pork production losses through reducing weight gain and causing respiratory disease in growing pigs.
Both viruses modulate the immune system, resulting in immunopathology of lymphocytes and macrophages. The resulting immunosuppression enhances the risk for primary and secondary bacterial and viral pathogens, and subsequently increases the need for administering antimicrobial treatments.
Since PRRSV was introduced over three decades ago, it is estimated to have cost at least $10 billion in losses to swine production. The most recent analysis of the cost of PRRS to the U.S. industry alone is approximately $664 million per year (Holtkamp et al., 2013). Economic losses are due to a decrease in production at the sow and growing pig levels, combined with increased costs of treatment and vaccination.
Although currently available PRRS modified live virus vaccines are widely used to reduce PRRS-associated production losses, they are generally thought inadequate for disease control and eradication (Vu et al., 2017). Programs designed for long-term PRRSV elimination from a herd are often unsuccessful, and PRRSV reintroduction is common.
Alternative strategies are urgently needed to control PRRS. The gut microbiome offers a nontraditional approach to improve weight gain and reduce morbidity and mortality associated with respiratory disease in the presence of PRRS.
Considering the potential impact of the gut microbiome on respiratory pathogens, our work has focused on identifying associations between the gut microbiota and disease outcome in nursery pigs under a PRRSV/PCV2 co-infection model. Our initial work demonstrated that characteristics of the gut microbiome were associated with clinical outcome of weaned pigs 70 days post-infection with PRRSV and PCV2 (Niederwerder et al., 2016).
Specifically, reduced clinical signs of PRRSV and PCV2, improved weight gain and reduced pulmonary pathology were associated with increased microbiome diversity and the presence of a nonpathogenic E. coli in the gut.
Following up this initial work, our second objective was to determine if preinfection gut microbiome characteristics were associated with subsequent outcome after co-infection with PRRSV and PCV2 in nursery pigs (Ober et al., 2017). Overall, pigs with high growth rates, reduced virus replication and less severe pneumonic lesions had several gut microbiome characteristics that may have predisposed or predicted outcome, including increased microbial diversity, reduced Methanobacteriaceae species, increased Ruminococcaceae species, and increased Streptococcaceae species.
Microbiome modulation
Once we established a relationship between gut microbiome characteristics and outcome following co-infection in our initial work, our third objective was to investigate microbiome modulation as a potential tool to improve respiratory disease outcome in nursery pigs.
Gut microbiome modulation was accomplished through fecal microbiota transplantation (FMT), which is the process by which feces are collected from a healthy donor and transplanted into a diseased or young individual. Similar to organ transplantation, the goal of FMT is to improve health or resolve disease in the transplant recipient.
To prepare the FMT for administration, donated feces are often mixed with sterile saline, blended to create a homogeneous mixture, passed through a filter to remove large fecal particulate, and processed to concentrate and protect the microorganisms prior to transplantation (Hamilton et al., 2012).
The FMT process not only transplants live and dead microbes, but also small feed particulate, cells from the small and large intestine, and metabolic products from microbes (Bojanova and Bordenstein, 2016). The mechanism by which FMT is efficacious is poorly understood for most diseases. However, the benefits derived from FMT are believed to be due to an increase in favorable microbes, restoration of normal flora, an increase in microbial diversity and stimulation of mucosal and systemic immunity (Niederwerder, 2018).
In humans, FMT is most commonly used as a therapeutic tool for recurrent Clostridium difficile infections unresponsive to antimicrobial treatment (Gough et al., 2011). However, FMT has recently been recognized as a potential therapy for a broad range of other diseases in humans, such as irritable bowel syndrome, multiple sclerosis, autism, chronic hepatitis B, antimicrobial resistant pathogen colonization, metabolic syndrome and insulin resistance (Bakker and Nieuwdorp, 2017; Cohen and Maharshak, 2017).
In animals, FMT — or transfaunation as it is described in ruminants — has also been used as a therapeutic tool for several diverse diseases, including use as part of a therapeutic protocol for canine parvovirus infection in dogs, left-displaced abomasum postsurgical complications in cattle, ulcerative colitis in cats, ruminal acidosis and atony in sheep, and equine acute colitis (Furmanski and Mor, 2017; Jasmin et al., 2011; Mullen et al., 2018; Pereira et al., 2018; Rager et al., 2004).
In addition to the therapeutic potential of FMT in animals, prophylactic and immunogenic uses of FMT have also been explored. For example, FMT has been investigated as a potential tool to increase feed efficiency in both poultry and swine (Niederwerder, 2018), and microbial feedback has been used as a tool to increase antibody production in sows and gilts specific to PEDV (Clement et al., 2016).
Tackling PRRSV, PCV2 with FMT
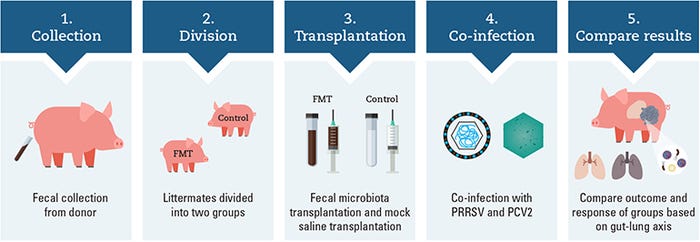
Work in our Kansas State University laboratory investigated the use of FMT as a method to modulate the microbiome for the prevention and control of PCV-associated disease and to mitigate the effects of PRRSV and PCV2 co-infection (Niederwerder et al., 2018). The illustration above outlines the overall experimental design investigating the potential role of the gut-lung axis in disease control of co-infected pigs.
Specifically, fecal microbes were collected from two donor sows with several characteristics of high health and productivity, including high parity; large litters; high percentage of piglets born alive; low preweaning mortality, with most mortalities associated with crushing injuries; a lack of fetal mummification in medical records; and no antimicrobial treatment received in the 15 months prior to donation.
Donated fecal material was screened and confirmed as negative for the presence of intestinal parasites. Feces were mixed with sterile saline, filtered and centrifuged to concentrate the microorganisms in the transplant material, and stored in 10% glycerol.
Ten pairs of barrow littermates were divided into FMT and control groups at weaning and balanced by arrival weight. The transplant material was delivered to weaned FMT piglets for seven consecutive days at a dose of 5 ml per day prior to co-infection. Control pigs received a sterile saline with glycerol mock-transplant for seven consecutive days at a dose of 5 ml per day prior to co-infection. All pigs were co-infected with PRRSV and PCV2 and followed for 42 days post infection.
Transplanted pigs were compared to saline mock-transplanted control pigs for clinical, virological and immunological outcomes. Pigs that received the FMT had reduced morbidity and mortality, decreased PRRSV and PCV2 replication, improved uniformity of weight gain, an enhanced immune response as demonstrated by increased antibody production, and a decreased requirement for antimicrobial administration.
Specifically, FMT prior to PRRSV and PCV2 co-infection was associated with a 70% reduction in mortality and a 60% reduction in the prescription of parenteral antimicrobial treatments. Finally, lung lesion severity was significantly reduced in pigs receiving the fecal transplantation.
This study provides evidence that microbiome modulation through FMT may be an alternative strategy in the preventive health care of pigs at risk for polymicrobial respiratory disease (Niederwerder et al., 2018).
Underestimated benefits
Prophylactic microbiome modulation and profiling the preinfection microbiome characteristics in swine provides a unique opportunity to consider possible avenues for how the microbiome may be used to prevent respiratory disease. Alternatives to antimicrobials and unconventional tools for disease control are essential for health care in food animal production.
The overarching theme of our work has consistently associated increased microbial diversity and beneficial microbe exposure with improved outcome after infectious respiratory challenges. Harnessing the beneficial characteristics of high-performing individuals through their gut microbiomes is a likely underestimated opportunity to increase herd health of swine (Niederwerder, 2018). Overall, gut microbes and FMT are exciting tools we may be able to use to provide respiratory relief for growing pigs.
Niederwerder is an assistant professor in the Department of Diagnostic Medicine/Pathobiology at the Kansas State University College of Veterinary Medicine.
References
Antunes, K.H., Fachi, J.L., de Paula, R., da Silva, E.F., Pral, L.P., Dos Santos, A.A., Dias, G.B.M., Vargas, J.E., Puga, R., Mayer, F.Q., Maito, F., Zarate-Blades, C.R., Ajami, N.J., Sant’Ana, M.R., Candreva, T., Rodrigues, H.G., Schmiele, M., Silva Clerici, M.T.P., Proenca-Modena, J.L., Vieira, A.T., Mackay, C.R., Mansur, D., Caballero, M.T., Marzec, J., Li, J., Wang, X., Bell, D., Polack, F.P., Kleeberger, S.R., Stein, R.T., Vinolo, M.A.R., de Souza, A.P.D., 2019. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nature communications 10, 3273.
Bakker, G.J., Nieuwdorp, M., 2017. Fecal Microbiota Transplantation: Therapeutic Potential for a Multitude of Diseases beyond Clostridium difficile. Microbiology spectrum 5.
Bojanova, D.P., Bordenstein, S.R., 2016. Fecal Transplants: What Is Being Transferred? PLoS biology 14, e1002503.
Clement, T., Singrey, A., Lawson, S., Okda, F., Nelson, J., Diel, D., Nelson, E.A., Christopher-Hennings, J., 2016. Measurement of neutralizing antibodies against porcine epidemic diarrhea virus in sow serum, colostrum, and milk samples and in piglet serum samples after feedback. Journal of Swine Health and Production 24, 147-153.
Cohen, N.A., Maharshak, N., 2017. Novel Indications for Fecal Microbial Transplantation: Update and Review of the Literature. Digestive diseases and sciences 62, 1131-1145.
Furmanski, S., Mor, T., 2017. First case report of fecal microbiota transplantation in a cat in Israel. Israel Journal of Veterinary Medicine 72, 35-41.
Gough, E., Shaikh, H., Manges, A.R., 2011. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 53, 994-1002.
Hamilton, M.J., Weingarden, A.R., Sadowsky, M.J., Khoruts, A., 2012. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. The American journal of gastroenterology 107, 761-767.
Holtkamp, D.J., Kliebenstein, J.B., Neumann, E.J., Zimmerman, J.J., Rotto, H.F., Yoder, T.K., Wang, C., Yeske, P.E., Mowrer, C.L., Haley, C.A., 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. Journal of Swine Health and Production 21, 72-84.
Jasmin, B.H., Boston, R.C., Modesto, R.B., Schaer, T.P., 2011. Perioperative ruminal pH changes in domestic sheep (Ovis aries) housed in a biomedical research setting. Journal of the American Association for Laboratory Animal Science : JAALAS 50, 27-32.
Messori, S., Trevisi, P., Simongiovanni, A., Priori, D., Bosi, P., 2013. Effect of susceptibility to enterotoxigenic Escherichia coli F4 and of dietary tryptophan on gut microbiota diversity observed in healthy young pigs. Veterinary microbiology 162, 173-179.
Mullen, K.R., Yasuda, K., Divers, T.J., Weese, J.S. 2018. Equine faecal microbiota transplant: Current knowledge, proposed guidelines and future directions, 151-160.
Niederwerder, M.C., 2017. Role of the microbiome in swine respiratory disease. Veterinary microbiology 209, 97-106.
Niederwerder, M.C., 2018. Fecal microbiota transplantation as a tool to treat and reduce susceptibility to disease in animals. Veterinary immunology and immunopathology 206, 65-72.
Niederwerder, M.C., Constance, L.A., Rowland, R.R.R., Abbas, W., Fernando, S.C., Potter, M.L., Sheahan, M.A., Burkey, T.E., Hesse, R.A., Cino-Ozuna, A.G., 2018. Fecal Microbiota Transplantation Is Associated With Reduced Morbidity and Mortality in Porcine Circovirus Associated Disease. Frontiers in microbiology 9, 1631.
Niederwerder, M.C., Jaing, C.J., Thissen, J.B., Cino-Ozuna, A.G., McLoughlin, K.S., Rowland, R.R.R., 2016. Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Veterinary microbiology 188, 1-11.
Ober, R.A., Thissen, J.B., Jaing, C.J., Cino-Ozuna, A.G., Rowland, R.R.R., Niederwerder, M.C., 2017. Increased microbiome diversity at the time of infection is associated with improved growth rates of pigs after co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2. Veterinary microbiology 208, 203-211.
Pereira, G.Q., Gomes, L.A., Santos, I.S., Alfieri, A.F., Weese, J.S., Costa, M.C., 2018. Fecal microbiota transplantation in puppies with canine parvovirus infection. Journal of veterinary internal medicine 32, 707-711.
Rager, K.D., George, L.W., House, J.K., DePeters, E.J., 2004. Evaluation of rumen transfaunation after surgical correction of left-sided displacement of the abomasum in cows. J Am Vet Med Assoc 225, 915-920.
Rosshart, S.P., Vassallo, B.G., Angeletti, D., Hutchinson, D.S., Morgan, A.P., Takeda, K., Hickman, H.D., McCulloch, J.A., Badger, J.H., Ajami, N.J., Trinchieri, G., Pardo-Manuel de Villena, F., Yewdell, J.W., Rehermann, B., 2017. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015-1028.e1013.
Schachtschneider, K.M., Yeoman, C.J., Isaacson, R.E., White, B.A., Schook, L.B., Pieters, M., 2013. Modulation of systemic immune responses through commensal gastrointestinal microbiota. PloS one 8, e53969.
Schuijt, T.J., Lankelma, J.M., Scicluna, B.P., de Sousa e Melo, F., Roelofs, J.J., de Boer, J.D., Hoogendijk, A.J., de Beer, R., de Vos, A., Belzer, C., de Vos, W.M., van der Poll, T., Wiersinga, W.J., 2016. The gut microbiota plays a protective role in the host defense against pneumococcal pneumonia. Gut 65, 575-583.
Sender, R., Fuchs, S., Milo, R., 2016. Revised Estimates for the Number of Human and Bacteria Cells in the Body.(Essay)(Report). PLoS biology 14, e1002533.
Song, D., Peng, Q., Chen, Y., Zhou, X., Zhang, F., Li, A., Huang, D., Wu, Q., Ye, Y., He, H., Wang, L., Tang, Y., 2017. Altered Gut Microbiota Profiles in Sows and Neonatal Piglets Associated with Porcine Epidemic Diarrhea Virus Infection. Scientific reports 7, 17439.
Vu, H.L.X., Pattnaik, A.K., Osorio, F.A., 2017. Strategies to broaden the cross-protective efficacy of vaccines against porcine reproductive and respiratory syndrome virus. Veterinary microbiology 206, 29-34.
Source: Megan C. Niederwerder, who is solely responsible for the information provided, and wholly owns the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like



