Tracing the origin of the novel PRRSV-2 L1C-1-4-4 variant
Most atypical losses from the Midwestern U.S. PRRS outbreaks in 2020-21 were caused by the same novel variant, L1C-1-4-4.
May 10, 2022

Currently, numerous breeding and growing pig herds in the Midwestern United States have suffered from massive porcine reproductive and respiratory syndrome outbreaks caused by the novel PRRSV-2 variant, L1C-1-4-4. Viruses belonging to this variant have highly similar genetic material, i.e. > 98% nucleotide identity in both open reading frame 5 (ORF5) gene and whole genome1. Although this variant is part of sub-lineage 1C (L1C)3, questions remain on how this variant rapidly spread and lead to atypical production losses.
Like other RNA viruses, the high mutation rate of PRRSV-2 allows it to adapt, survive and sometimes become more virulent. Previous studies demonstrate that virulence and antigenicity of PRRSV-2 is determined by several regions throughout the genome5,6. At the same time, most efforts to analyze PRRSV-2 sequences focus only the ORF5 gene, which represents just 4% of the genome. Thus, analyzing whole genome sequences (WGSs) of the L1C-1-4-4 variant is a first crucial step to understand how this virus differs from other circulating variants.
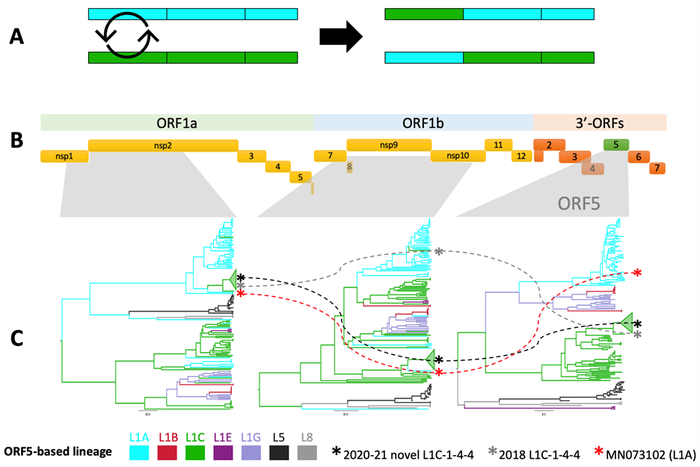
Inferring a virus's evolutionary origin from whole genomes is complicated due to the occurrence of recombination, which is an evolutionary mechanism by which genomic fragments are exchanged between viruses (Figure 1A). Such events contribute to the genetic diversity in the virus population and potentially lead to rapid changes in a virus's genetic makeup and, potentially, its phenotype. Moreover, recombination can result in distinct evolutionary histories amongst different recombined genomic fragments, which makes WGS-based analysis hard to interpret.
We screened a set of PRRSV-2 whole genomes for recombination, including the novel L1C-1-4-4 variant (n = 19) and other isolates collected in the United States over the past two decades (n = 232). Sequences were classified into lineages (L1-9) and sub-lineages (L1A-H) according to shared ancestry in the ORF5 gene. Three distinct WGS fragments were then used to estimate the virus's evolutionary history and frequency of ancestral inter-(sub)lineage recombination.
The analyses confirmed that the novel L1C-1-4-4 variant was a recombinant virus with a L1C-like genomic backbone. The variant diverged in late 2018 to early 2019, acquiring a non-structural protein 2 coding region from L1A viruses (RFLP 1-7-4) through recombination. The closest relatives were two isolates belonging to L1C and L1A collected in 2018, both of which had a different recombination history than the novel variant (Figure 1C). However, our analysis suggests that inter-lineage recombination events resulting in widespread transmission (i.e., those that leave detectable numbers of descendants) were relatively uncommon (< 0.5 events/year).
Given these results coupled with the field epidemiological situation, rapid exchange of genetic material through recombination between co-circulating PRRSV-2 strains may be a possible mechanism associated with variant emergence. While recombination may have contributed to the recent epidemic caused by the novel L1C-1-4-4 variant, recombination events that leave discernable traces on PRRSV-2 trees appear somewhat rare. However, co-circulation of multiple, genetically-diverse lineages likely increases the chances that recombination will occasionally result in novel variants that harbor combinations of traits that increase their capabilities to infect and spread in host populations. Further studies on an impact of this recombination on PRRSV-2 virulence are needed.
In summary, here are key observations about the novel PRRSV-2 L1C-1-4-4 variant:
Most atypical losses from the Midwestern U.S. PRRS outbreaks in 2020-21 were caused by the same novel variant, L1C-1-4-4.
The novel variant is a recombinant virus with a L1C-like genomic backbone.
The variant diverged from other circulating PRRSV-2 viruses in late 2018- 2019, around which time it acquired a partial nsp2 coding region from L1A-like viruses (the sub-lineage that includes RFLP 1-7-4) via recombination.
It is currently unknown if or how recombination effected the phenotype of the virus.
Viruses belonging to the same ORF5-based lineage largely remain clustered together in other parts of the genome, though some evidence of recombination.
This research was funded by the joint NIFA-NSF-NIH Ecology and Evolution of Infectious Disease award 2019-67015-29918, the University of Minnesota College of Veterinary Medicine Signature Programs, Grant Number MIN-62-133, and the Swine Health Information Center.
Source: Nakarin Pamornchainavakul, Mariana Kikuti, Igor A. D. Paploski, Dennis N. Makau, Albert Rovira, Cesar A. Corzo and Kimberly VanderWaal, who are solely responsible for the information provided, and wholly own the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
References
1. Kikuti, M. et al. Emergence of a New Lineage 1C Variant of Porcine Reproductive and Respiratory Syndrome Virus 2 in the United States. Front. Vet. Sci. (2021). doi:10.3389/fvets.2021.752938
2. Wissink, E. H. J. et al. The major envelope protein, GP5, of a European porcine reproductive and respiratory syndrome virus contains a neutralization epitope in its N-terminal ectodomain. J. Gen. Virol. (2003). doi:10.1099/vir.0.18957-0
3. Paploski, I. A. D. et al. Phylogenetic structure and sequential dominance of sub-lineages of prrsv type-2 lineage 1 in the United States. Vaccines (2021). doi:10.3390/vaccines9060608
4. Snijder, E. J., Kikkert, M. & Fang, Y. Arterivirus molecular biology and pathogenesis. Journal of General Virology (2013). doi:10.1099/vir.0.056341-0
5. Kwon, B., Ansari, I. H., Pattnaik, A. K. & Osorio, F. A. Identification of virulence determinants of porcine reproductive and respiratory syndrome virus through construction of chimeric clones. Virology (2008). doi:10.1016/j.virol.2008.07.030
6. Ruedas-Torres, I. et al. The jigsaw of PRRSV virulence. Veterinary Microbiology (2021). doi:10.1016/j.vetmic.2021.109168
7. Simon-Loriere, E. & Holmes, E. C. Why do RNA viruses recombine? Nature Reviews Microbiology (2011). doi:10.1038/nrmicro2614
8. Martin, D. P. et al. RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. (2021). doi:10.1093/ve/veaa087
You May Also Like



