Sequencing technologies improve swine infectious disease management
Infectious disease diagnostics using MinION sequencing in a laboratory setting has been proven to be feasible, which can be used as a support for disease investigation.
April 14, 2020

Swine infectious disease is a major threat to the swine industry, causing great economic losses and challenges stable pork production. The development of rapid and accurate diagnostic methods is important for effective infectious disease control.
Sequencing technologies, including Sanger sequencing and Illumina sequencing, have been supporting the control of swine infectious disease for several decades. In recent years, the long-read Oxford Nanopore MinION sequencing technology has gained much attention due to its unique features of high accessibility, portability, easy operation and real-time data streaming and analysis. MinION sequencing has the advantage of needing no prior sequence information about a pathogen unlike Sanger sequencing and has advantages over Illumina sequencing in terms of accessibility and infrastructure cost. In addition, MinION sequencing provides a new solution for on-site, real-time disease investigation, which needed scientific evaluation of its performance for infectious disease control, as well as validation and standardization of the sequencing method for use in clinical cases.
Since 2015, we have been developing novel diagnostic methods that use Oxford Nanopore MinION sequencing and bioinformatics tools to aid in investigation of swine bacterial (using Streptococcus suis as a model) and viral diseases (using porcine reproductive and respiratory syndrome virus and Senecavirus A as models). We have developed detailed guidelines for the relevant wet-lab operation (such as DNA/RNA extraction, quality control and sequencing library preparation) and dry-lab bioinformatics analysis (such as assembly, genotyping and taxonomic analysis).
In order to improve accuracy of MinION sequencing for bacterial functional genomic characterization, de novo assembly was performed to generate a whole genome consensus, which was then followed by antibiotic resistance gene annotation. We determined that MinION sequencing can be used to predict antibiotic resistance based on gene presence, allowing for guidance of effective antibiotic use.
Secondly, using PRRSV as an example, we demonstrated that MinION sequencing was able to detect and identify virus at the strain level, allowing for precise investigation of outbreaks that occur in vaccinated herds or co-infection of multiple field-strains. Also, a whole genome viral sequence was able to be generated, which can help further our understanding of viral evolution, infection disease epidemiology and pathogenic mechanisms.
In addition, using Senecavirus A as a model, we demonstrated that MinION sequencing could be used for rapid investigation of emerging pathogens by species, strain and whole genome sequence. The pipeline we developed is robust and unbiased, it doesn't need prior sequence information about the pathogens under detection. Of note, our whole workflow starting from receiving a biological sample until identification of the viral strain present took less than a day, allowing for rapid, same-day report generation (Figure 1).
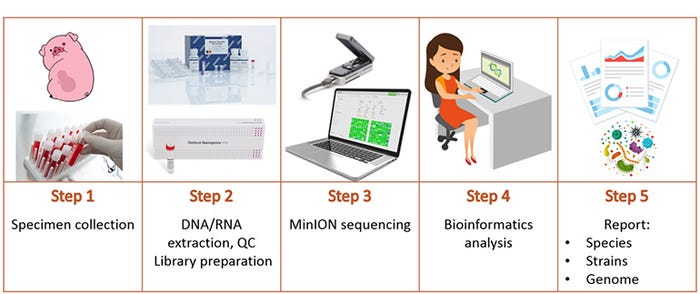
Our studies provide hands-on references for bacterial and viral disease characterization using sequencing and bioinformatics tools, which highlight the potential of the portable MinION sequencer for swine infectious disease management. At the same time, limitations and future directions are acknowledged.
Major limitations that need to be addressed include difficulty of on-site extraction and relatively low analytical sensitivity of sequencing. New extraction kits and library preparation devices designed for field work have been developed to enable on-site diagnosis and need to be examined and optimized. More importantly, additional research is needed to improve the detection limit of sequencing to allow for examination of samples with low infection levels.
Today, infectious disease diagnostics using MinION sequencing in a laboratory setting has been proven to be feasible, which can be used as a support for disease investigation. Going forward, as technology develops it is only a matter of time before MinION portable sequencing can be used as a routine diagnostic tool for outbreak investigations and disease surveillance to provide real-time insights.
Acknowledgements
Michael P. Murtaugh, who passed away Sept. 18, 2018, contributed to this work.
The research was supported by grant funding from Boehringer Ingelheim and the National Pork Board, and fellowships to Shaoyuan Tan from the Leman China Conference and MnDRIVE Global Food Ventures.
Tan S., Dvorak CMT, Estrada AA, Gebhart C, Marthaler DG, Murtaugh MP. MinION sequencing of Streptococcus suis allows for functional characterization of bacteria by multilocus sequence typing and antimicrobial resistance profiling. J Microbiol Methods. 2020 Feb;169:105817. doi: 10.1016/j.mimet.2019.105817.
Tan S., Dvorak CMT, Murtaugh MP. Rapid, Unbiased PRRSV Strain Detection Using MinION Direct RNA Sequencing and Bioinformatics Tools. Viruses. 2019 Dec 7;11(12). pii: E1132. doi: 10.3390/v11121132.
Tan S., Dvorak CMT, Murtaugh MP. Characterization of emerging swine viral disease through Oxford Nanopore sequencing using Senecavirus A as a model. In progress.
Sources: Shaoyuan Tan and Cheryl M.T. Dvorak, who are solely responsible for the information provided, and wholly own the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like



