PRRSV detection dynamics in U.S.
Blueprint: Differences between Midwest, Southeast exist.
October 30, 2019

By Cesar Moura, Gustavo Silva and Daniel Linhares, Iowa State University; Reid Philips and Eduardo Fano, Boehringer Ingelheim Animal Health
Porcine reproductive and respiratory syndrome virus causes a considerable economic impact on the swine industry worldwide by increasing reproduction losses and respiratory disorders. Fifty-five percent of the economic impact is on the growing Phase 1.
Therefore, there is a critical need to better understand how the virus spreads within infected populations of growing pigs in the United States and how it relates to growing pig performance, under field conditions.
We conducted a series of field studies with the objectives to:
identify patterns of wild-type PRRSV-1 and PRRSV-2 detections over time of vaccinated growing pig populations in the United States.
assess the effect of the different wild-type PRRSV-1 and PRRSV-2 detection patterns on mortality of vaccinated growing pig populations from the United States.
assess the effect of wild-type PRRSV-2 detection frequency and wild-type PRRSV-2 diversity on the mortality of vaccinated growing pig populations from the United States.
Methods
Eighty-one vaccinated growing pig groups from the Southeast or Midwest regions of the United States were monitored during the wean-to-finish phase, by collecting six spatially distributed1 oral fluids samples every three weeks.
Samples were collected at eight time points (4, 7, 10, 13, 16, 19, 22 and 25 weeks of age). All samples were submitted to the Iowa State University Veterinary Diagnostic Laboratory for detection of PRRSV RNA by RT-qPCR.
When there was at least one sample testing PCR-positive for PRRSV-2 (per time point), the one with lowest cycle threshold value was submitted to open reading frame-5 sequencing to differentiate between vaccine-like, or wild-type, PRRSV. All positive samples for PRRSV-1 were assumed to be wild-type virus, since there was no vaccine against this genotype used in the study population. All pig groups were:
originated from PRRSV-positive stable or unstable sow farms (based on the 2011 American Association of Swine Veterinarians PRRSV classification2)
previously vaccinated with PRRSV-2 modified live virus vaccine at the sow farm
placed at areas highly dense with pigs
Statistical analyses were performed using the growing pig group as the experimental unit.
Clustering analysis was performed using K-means methods on R program, to describe different patterns of wild-type PRRSV-1 and PRRSV-2 detections over time from study groups.
Hierarchical generalized linear mixed regression models, on an SAS program, were built to compare differences of growing pig mortality among clusters, and to measure the effect of the wild-type PRRSV-2 detection frequency and wild-type PRRSV-2 diversity on growing pig mortality.
Results
The study included 119,492 growing pigs from 81 groups, with 648 sampling time points totaling 3,888 oral fluids samples.
PRRSV-2 dynamics. We detected wild-type PRRSV-2 in 74 of 81 (91.4%) growing pig groups included on the study at least once during the wean-to-finish phase.
Three different clusters of wild-type PRRSV-2 infection dynamics were generated, using the K-means clustering method (Figure 1, dotted lines). These different infection dynamics represented an early-infection cluster as a blue line (n=33); a mid-infection cluster as a tan line (n=30); and a late-infection cluster as a brown line (n=11).
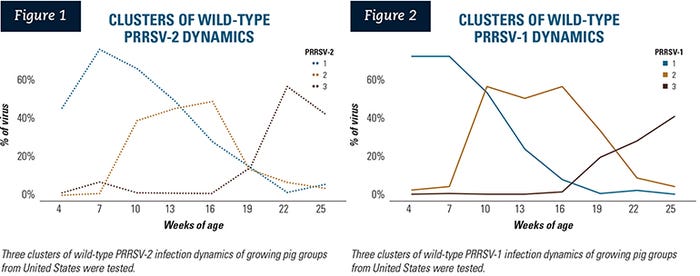
Seven groups had no wild-type PRRSV-2 detection during the wean-to-finish phase and therefore were not included in the clustering analysis. These pig groups were classified as having no PRRSV-2 detection, and were defined as the control group for the regression analyses.
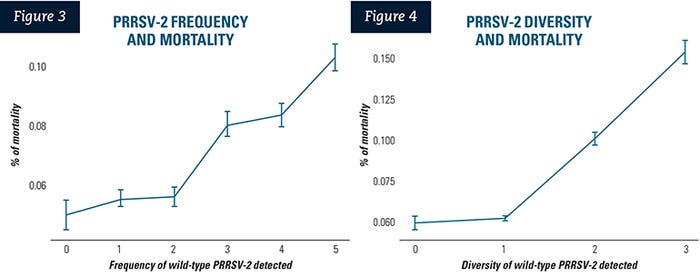
The early-infection cluster had the highest growing pig mortality rate, as in Table 1: (10.86% [95% confidence interval 7.85% to 15.01%]) followed by the mid-infection cluster (9.18% [95% CI 6.62% to 12.75%]), the late-infection cluster (8.44% [95% CI 6.03% to 11.8%]) and the non-PRRSV-detection cluster (7.92% [95% CI 5.61% to 11.18%]).
Both early and mid-infection clusters had significantly higher growing pig mortality, when compared to the mortality rate of the control group (p<0.0001 and p=0.0082, respectively). The growing pig mortality of the late-infection cluster was not statistically different from the control group (p=0.3712) (Table 1).
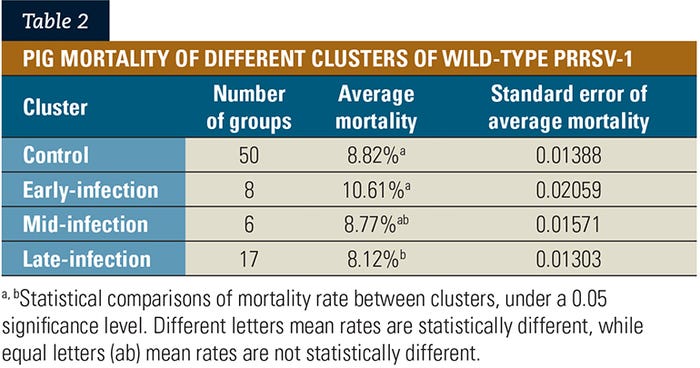
As the wild-type PRRSV-2 detection frequency (Figure 3) and the wild-type PRRSV-2 diversity (Figure 4) increased, the growing pig mortality also increased. It shows that the higher the PRRSV-2 challenge on the growing pig groups, the higher the growing pig mortality.
In this situation, challenge was measured by the number of times that each group was detected with wild-type PRRSV-2 (detection frequency; see Figure 3), and the number of genetically different wild-type PRRSV-2 was detected in each group (wild-type PRRSV diversity; see Figure 4). Virus genetic composition was based on restriction fragment-length polymorphism patterns of the ORF-5 sequencing.
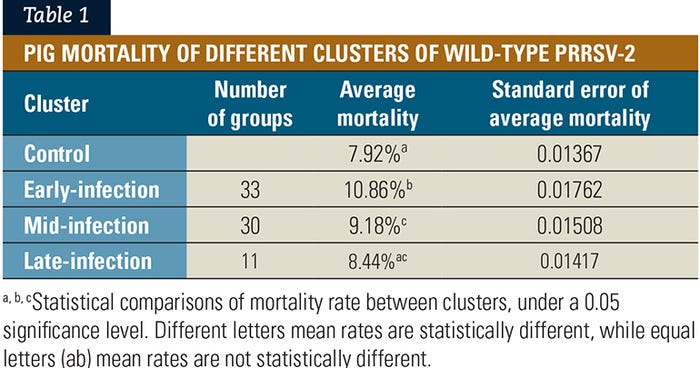
PRRSV-1 dynamics. We detected wild-type PRRSV-1 in 31 of 81 (38.3%) growing pig groups at least once in the wean-to-finish phase.
Three different clusters of wild-type PRRSV-1 infection dynamics were also generated, using the K-means clustering method (see Figure 2). These different infection dynamics represent an early-infection cluster as a blue line (n=8); a mid-infection cluster as a tan line (n=6); and a late-infection cluster as a brown line (n=17).
Fifty additional groups had no wild-type PRRSV-1 detection during the wean-to-finish phase and were not included on the clustering analysis. These pig groups were classified as having no PRRSV-1 detection and were defined as the control group.
The early-infection cluster had the highest growing pig mortality rate as in Table 2: (10.61% [95% CI 7.20% to 15.63%]) followed by the mid-infection cluster (8.77% [95% CI 6.14% to 12.54%]) and the late-infection cluster (8.12% [95% CI 5.90% to 11.19%]).
The non PRRSV-1 detection cluster had an average of 8.82% in the growing pig mortality (95% CI 6.44% - 12.07%). The early-infection cluster had significantly higher growing pig mortality, when compared to the mortality rate of the late-infection cluster (p=0.0153).
Growing pig mortality of the late-infection cluster was significantly lower when compared to the control group (p=0.0327) (Table 2).
Discussion
Overall, 92% of study groups were detected with wild-type PRRSV type 2 at least once during the wean-to-finish phase. On the groups raised in the Southeast region of the United States, the percentage of positive groups was 89%; while on the groups raised in the Midwest, it was 96%.
Only 38% of study groups were detected with wild-type PRRSV type 1 at least once during the wean-to-finish phase. Interestingly, all the wild-type PRRSV-1 positive groups belonged to the Southeast region, and there was no PRRSV-1 detection in the Midwest region.
Results suggest that vaccinated growing pigs raised in areas of high pig density in the Southeast or Midwest regions of the United States are likely to be exposed to wild-type PRRSV-2 during the wean-to-finish period. The detection of the American genotype virus was strongly associated with increased mortality. Moreover, the earlier the virus detection, the higher the subsequent mortality rate.
The RFLP patterns in figures 3 and 4 were used for measuring virus genetic background; however, this is one of the limitations of the study. Interpretations of these patterns must be done with caution, since there is significant genetic variation within RFLP types of wild-type PRRSV.
Under the study conditions, more than half (56%) of the groups raised in the Southeast were exposed to PRRSV-1, while none of the groups in the Midwest were exposed to this genotype. It was not possible to associate wild-type PRRSV-1 detection to increased growing pig mortality.
Moura, Silva and Linhares work in the Iowa State University Veterinary Diagnostic and Production Animal Medicine Department. Philips is a senior technical manager and Fano a technical manager, both with Boehringer Ingelheim Animal Health.
References
1. Rotolo ML, Sun Y, Wang C, et al. Sampling guidelines for oral fluid-based surveys of group-housed animals. Vet Microbiol. 2017;209:20-29.
2. Holtkamp D, Polson D, Torremorell M. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. Journal of Swine Health and Production. 2011;19(1):44-56.
Sources: Cesar Moura, Gustavo Silva, Daniel Linhares, Reid Philips and Eduardo Fano, who are solely responsible for the information provided, and wholly own the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like


