Practical use of pig movement data for disease interventions
Understanding stability of movement networks through time can inform decisions for designing, implementing disease management.
July 13, 2021

The transport of animals plays a significant role in infectious disease transmission between farms [1]–[4], resulting in a highly inter-connected web of connections amongst farms that make the swine industry vulnerable to disease epidemics. An understanding of these animal movement networks is crucial when evaluating risk of disease spread at the national, regional, or farm-level and informing decisions on disease mitigation and prevention. In addition, evaluating network connections also creates opportunities for strategic interventions that target potential super-spreaders and fragment infection chains in the network. The size of outbreaks can be minimized, and emerging diseases can be detected earlier by identifying and targeting control and surveillance efforts towards key, highly connected farms, thus reducing the spread of a pathogen and associated economic losses.
However, to effectively utilize animal movement data for disease control, pertinent questions need to be addressed about how many months of data are needed to obtain a stable and complete picture of a movement network, how long the data remains relevant after collection, and how often we need to update analyses of network connections. The absence of such information constrains the deployment of network-based targeted control strategies in swine production systems. To help answer these questions for the U.S. swine industry, we used network theory to analyze 282,807 pig movements among 2,724 farms from two production systems in the U.S. between 2014 and 2017.
We observed that targeting interventions to highly connected farms (i.e., farms with high “degree” in the network) was far more effective at fragmenting potential transmission chains than random interventions (Figure 1). Moreover, using pig movement data as old as two years to identify and target high-risk farms was as effective at fragmenting the current network as utilizing current data; the effectiveness/reliability decreased considerably once the data was more than 30 months old.
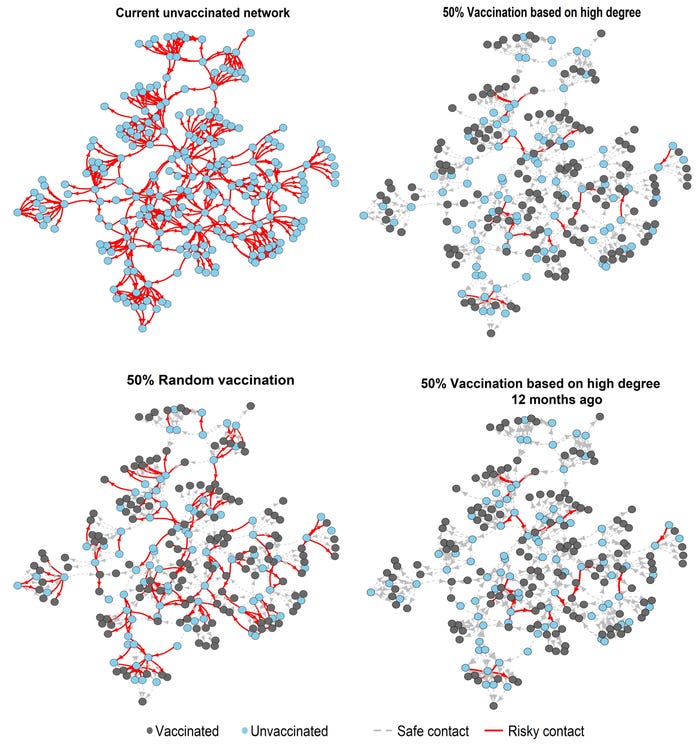
Using network simulations, we also observed that vaccinating 10-15% of highly connected farms effectively disrupted disease transmission through the network more broadly (assuming 100% vaccine effectiveness and disease transmission primarily through animal movements). In contrast, if farms were randomly selected for vaccination, more than 60% of the farms would have to be vaccinated to achieve a similar impact.
For practical implementation of such network-based interventions, we show that using animal movement data aggregated over time windows of six months provides a stable picture of farm-connectivity networks and allows for reliable identification of farms that may function as super-spreaders. Targeted interventions to these farms can effectively disrupt disease transmission.
The temporal stability of the networks was due to the repeatability of specific between-farm pig shipments (loyalty). About 50 – 60% of specific weaned pig movements were repeated at least once within a 52-week interval, while 12 – 22% of specific feeder pig movements were repeated at least once in the same interval. Although overall loyalty decreased through time, we observed cyclic episodes of increased loyalty every eight and 17-20 weeks for weaned and feeder pigs, respectively. These cycles corresponded to the pig lifecycles and expected time spent at the two stages of production in a multisite production system.
In summary, understanding the stability of movement networks through time can inform decisions for designing and implementing disease management, monitoring and surveillance interventions. Six months of data is suggested for selecting farms for targeted interventions. Historical movement data (up to 24 months old) is almost as effective as up-to-date data for identifying highly-connected farms, and both perform better than randomly deployed interventions. Thus, disease mitigation measures targeted to 10-15% highly connected farms/ super-spreaders can be cost-effective and efficient for disease management. Strong loyalty between farms may be beneficial in reducing disease spread due to repeated contacts between fewer farms. While changes in loyalty patterns may occur due to major disturbances in population structure and connectivity, understanding baseline loyalty is beneficial as a benchmark against which we can evaluate changes in movement patterns.
This work has been published in: D. N. Makau, I. A. D. Paploski, and K. VanderWaal, “Temporal stability of swine movement networks in the U.S.,” Prev. Vet. Med., vol. 191, 2021. https://doi.org/10.1016/j.prevetmed.2021.105369.
Source: By Dennis Makau, Igor Paploski, Kimberly VanderWaal, University of Minnesota, who are solely responsible for the information provided, and wholly owns the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset. The opinions of this writer are not necessarily those of Farm Progress/Informa.
References
1. A. C. Kinsley, A. M. Perez, M. E. Craft, and K. L. Vanderwaal, “Characterization of swine movements in the United States and implications for disease control,” Prev. Vet. Med., vol. 164, no. April 2018, pp. 1–9, Mar. 2019.
2. K. VanderWaal, A. Perez, M. Torremorrell, R. M. Morrison, and M. Craft, “Role of animal movement and indirect contact among farms in transmission of porcine epidemic diarrhea virus,” Epidemics, vol. 24, no. April 2018, pp. 67–75, 2018.
3. K. VanderWaal, I. A. D. Paploski, D. N. Makau, and C. A. Corzo, “Contrasting animal movement and spatial connectivity networks in shaping transmission pathways of a genetically diverse virus,” Prev. Vet. Med., vol. 178, no. March, p. 104977, May 2020.
4. D. N. Makau, I. A. D. Paploski, C. A. Corzo, and K. VanderWaal, “Dynamic network connectivity influences the spread of a sub‐lineage of porcine reproductive and respiratory syndrome virus,” Transbound. Emerg. Dis., vol. 00, pp. 1–14, Feb. 2021.
5. D. N. Makau, I. A. D. Paploski, and K. VanderWaal, “Temporal stability of swine movement networks in the U.S.,” Prev. Vet. Med., vol. 191, 2021.
You May Also Like



