Monitoring PRRS virus at low prevalence
Blueprint: Methods to monitor PRRS virus in breeding herds based on population sampling can be used and are commercially available.
October 30, 2018

By Marcelo Nunes de Almeida, Gustavo de Sousa e Silva, Alejandro Ramirez, Derald Holtkamp, Chris Rademacher, Locke Karriker and Daniel Linhares, Iowa State University
Porcine reproductive and respiratory syndrome is caused by the PRRS virus, an RNA organism with a relatively high mutation rate. PRRS costs the swine industry in the United States $664 million annually (Holtkamp et al., 2013). Therefore, adoption of prevention, detection, and control or elimination practices are of extreme importance in regions and/or countries where the virus is endemic.
PRRS virus infection results in viremia for a period of approximately three to five weeks (detected by polymerase chain reaction in blood serum) (Xiao et al., 2004), followed by a period of viral shedding (measured by detection in oral fluids using PCR) up to 90 days postinfection (Batista et al., 2004). Seroconversion occurs relatively fast, where 90% of animals seroconvert within seven to 14 days postinfection (detected by ELISA in serum, or oral fluids) (Prickett et al., 2008a; Prickett et al., 2008b).
Clinical signs are a result of a systemic infection associated with lack of appetite (first days after infection as a result of the fever), abortions, early farrows, reproductive failures (including sow abortions and boar infertility), and significant decrease in growth performance (increased mortality, decreased feed efficiency and lower average daily weight gain) (Rossow et al., 1994; Halbur et al., 1995; Halbur et al., 1996; Rossow, 1998; Rossow et al., 1999).
The confirmation of PRRS outbreaks can be done through the combination of laboratory tests, epidemiological information and clinical examination. Among the laboratory techniques, molecular (PRRS virus RNA detection using PCR) and antibody (detection using ELISA) tests are the most commonly used to detect PRRS virus exposure. Current protocols include the use of 30 serum samples from due-to-wean piglets to detect PRRS virus in breeding herds (Holtkamp et al., 2011).
One limitation of this protocol is that it targets the detection of the virus when the prevalence is 10% or higher. For situations when PRRS virus is circulating with a lower prevalence than 10%, this protocol is likely to miss the detection of the virus.
The need for improved PRRS virus detection, especially at low prevalence, has led to the recent development of new tools to surveil for the virus in large operations. These new monitoring and surveillance methods need to be sensitive, specific and fast. This information will focus on the detection of PRRS virus at the population level. First, we will discuss processing fluids (exudate from testicles and tails) obtained from 3- to 5-day-old piglets. Second, we will describe the detection of PRRS virus using family oral fluids obtained from due-to-wean piglets. Finally, we will discuss the potential for monitoring production parameters to assist detection of possible PRRS outbreaks in breeding herds.
Processing fluids
PRRS virus-monitoring based on molecular (PCR) and/or serologic (ELISA) tests in processing fluids was recently reported by Lopez et al., 2018, and its use has been increasing in the U.S. Processing fluids consist of an exudate from the tail docking and castration of nursing piglets (usually between 3 and 5 days old).
It consists of placing the docked tails and testicles in a container (often a bucket), where a disposable plastic bag is placed and covered with cheesecloth or other type of filter to strain tissues (see photos).
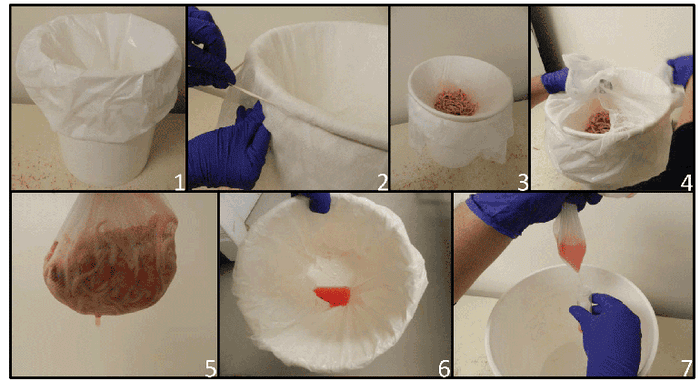
Here are the steps for collecting processing fluids: 1. Plastic bag is put into clean bucket. 2. Cheesecloth is placed over mouth of bucket to hold tissues and allow fluid to pass into plastic bag. 3. Tissues are placed on cheesecloth. 4 and 5. Tissues are removed after collection. 6. Processing fluid is recovered in plastic bag. 7. Fluid decanted from plastic bag into tube.
Serosanguineous fluids will progressively be filtered into the plastic bag. Theses fluids can then be transferred to a 50-milliliter tube and sent to a laboratory. Results from several field trials have shown improved sensitivity and specificity of processing fluids compared to 30 or 60 individual serum samples from piglets when used to determine if a farm or group of animals is positive or negative for PRRS virus (Lopez et al., 2017).
Part of the improved sensitivity of processing fluids (compared to individual samples) is in the fact that the sample represents hundreds of animals — in theory, all piglets being “processed” release fluids from the cut surfaces of the tail and/or testicles, which can be used for testing. Collecting processing fluids can be executed in a simple, practical and fast way by farm staff, which contributes to rapid adoption of the technique for routine monitoring.
The advantages of processing fluids-based monitoring include the feasibility of testing many more pigs more frequently, and at a lower cost. Farms can collect and test samples using processing fluids on a weekly or daily basis, compared to the usual monthly individual serum samples used.
Processing fluid-based PRRS virus monitoring is also significantly more economical than the conventional methods (e.g., 30 to 60 serum samples). As an example, in a farrowing room with 38 crates, a sample of processing fluids representing approximately 500 pigs would cost one PCR test ($25), or 5 cents per piglet.
In comparison, to monitor the same population using individual serum samples, 60 piglets would be needed, allowing a 95% confidence to detect PRRS virus with a 5% prevalence or higher. Those samples could be tested in pools of 1:5, for a total of 12 PCRs or 60 cents per piglet.
Family oral fluids
The literature on the use of oral fluids in due-to-wean piglets is sparse. This is due to the fact that obtaining oral fluids from this population is not as straightforward as it is for the growing or reproductive-age population. Young piglets are not easily attracted to chew on ropes, making it difficult to collect oral fluids.
Thus, an alternative approach to collect oral fluids from due-to-wean piglets has been reported and called family oral fluids (Yeske-Livermore et al., 2014; Almeida et al., 2017; Almeida et al., 2018a; Almeida et al., 2018b).
FOF consists of hanging a cotton rope in the front of the crate, where both the sow and piglets have access to it. The sow will usually chew on the rope and piglets typically mimic her behavior, allowing for the collection of oral fluids from the due-to-wean population (see photo at the top).
Almeida and others showed that the success rate of collecting FOF has been greater than 90%, while collecting oral fluids by hanging a rope in the back of the crate where only the piglets had access to it (traditional-litter oral fluids) had only a 43.5% success rate (Almeida et al., 2018b).
In another experiment, PRRS virus detection in individual piglet serum versus FOF was compared in 72 litters (718 piglets). Serum samples were collected from all piglets in litters where oral fluids were collected. Samples were tested by PCR (Almeida et al., 2018a). In serum, 24.2% (174 of 718) of samples were positive. Thirty-two litters (44.4%) had greater than or equal to one piglet positive for PRRS virus.
Among these 32 litters, 27 (84.4%) of the FOF were PCR-positive. All litters with greater than or equal to three viremic piglets (greater than or equal to three PCR-positive samples in a litter) were positive on FOF, compared to 50% of litters (5/10) with one or two viremic piglets. All litters with negative serum results (n = 40) were also negative by FOF, indicating excellent test diagnostic specificity. Family oral fluids were able to detect 87.5% of positive individual samples (sensitivity).
Family oral fluids are a combination of sow, piglet and perhaps environmental sampling. The combined results described above support that FOF-based sampling is a practical and effective way to monitor PRRS virus in due-to-wean litters.
Production parameters
PRRS virus outbreaks in breeding herds usually result in significant changes in a few production parameters, like the increase in the number of abortions, neonatal losses (stillborn and mummified piglets) and preweaning mortality.
Based on these changes, Gustavo Silva and others reported the use of production data to monitor the detection of PRRS outbreaks in breeding herds (Silva et al., 2017). More recently, Cesar Moura and collaborators developed an online application to collect production data and assess deviations in productivity based in the algorithms previously validated (Silva et al., 2018).
In summary, the application offers resources to capture data on a daily or weekly basis of key production parameters, including the number of sows off feed in gestation, the number of abortions, the number of neonatal losses and preweaning mortality. A statistical process control screening is performed and checked for significant deviations on productivity.
When a deviation is detected, an automated email is sent to specific users (sow farm manager and/or veterinarian) alerting them of a possible outbreak, who can then trigger the collection of samples for tests.
The automated monitoring of production data is a simple, low-cost and efficient strategy that can serve as a screening process for farms, and should always be followed with laboratory monitoring to investigate causes of deviation in productivity.
Conclusion
In summary, there are several ways to monitor herds for PRRS, including the use of laboratory tests and/or production data. Conventional methods of monitoring are based on testing a significant number of individual samples, which take time to be collected and processed, may cause logistical complications and demand significant investment in laboratory tests.
Alternatively, methods to monitor PRRS virus in breeding herds based on population sampling (processing fluids, family oral fluids and production parameter monitoring) can be used and are commercially available. They allow for a larger number of animals to be sampled and higher herd sensitivity at lower cost and in a more practical way. The new approaches can make detecting PRRS virus, especially at low prevalence (below 10%), practical and more feasible.
References
Almeida, M.N., Philips, R., Linhares, D.C.L., 2017. PRRSV surveillance on the breeding herd — is oral fluids from suckling piglets an alternative?, pig333.com/prrs/prrsv-surveillance-on-the-breeding-herd-is-oral-fluids-from-suckling_12334/.
Almeida, M.N., Zimmerman, J.J., Holtkamp, D.J., Rademacher, C., Rotto, H., Schneider, P., Linhares, D.C.L., 2018a. Comparative evaluation of family oral fluids and piglet sera to detect PRRSV RNA by PCR. In: Veterinarians, A.-A.A.o.S. (Ed.), Annual Meeting of the American Association of Swine Veterinarians, San Diego.
Almeida, M.N.Z., J.J., Holtkamp, D.J., Rademacher, C., Rotto, H., Schneider, P., Linhares, D.C.L., 2018b. Detecting PRRSV at low prevalence in due-to-wean piglets using oral fluids. In: AASV (Ed.), Annual Meeting of the American Association of Swine Veterinarians. San Diego.
Batista, L., Pijoan, C., Dee, S., Olin, M., Molitor, T., Joo, H.S., Xiao, Z., Murtaugh, M., 2004. Virological and immunological responses to porcine reproductive and respiratory syndrome virus in a large population of gilts. Canadian journal of veterinary research 68, 267-273.
Halbur, P.G., Paul, P.S., Frey, M.L., Landgraf, J., Eernisse, K., Meng, X.J., Andrews, J.J., Lum, M.A., Rathje, J.A., 1996. Comparison of the antigen distribution of two U.S. porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 33, 159-170.
Halbur, P.G., Paul, P.S., Frey, M.L., Landgraf, J., Eernisse, K., Meng, X.J., Lum, M.A., Andrews, J.J., Rathje, J.A., 1995. Comparison of the pathogenicity of two U.S. porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol 32, 648-660.
Holtkamp, D., Kliebenstein, J., Neumann, E., Zimmerman, J., Rotto, H., Yoder, T., Wang, C., Yeske, P., Mowrer, C., Haley, C., 2013. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. Journal of Swine Health and Production, 72-84.
Holtkamp, D., Polson, D., Torremorell, M., Morrison, R., Classen, D., Becton, L., Henry, S., Rodibaugh, M.T., Rowland, R.R., Snelson, H., Straw, B., Yeske, P., Zimmerman, J., 2011. Terminology for classifying swine herds by porcine reproductive and respiratory syndrome virus status. J Swine Health Prod 19, 44-56.
Lopez, W., Angulo, J., Zimmerman, J., Linhares, D., 2017. Processing fluids, blood serum, and tail blood swabs to detect PRRSV RNA and PCV2 DNA by PCR-based assays. In, James McKean Swine Disease Conference, Ames, Iowa, 65.
Lopez, W.A., Angulo, J., Zimmerman, J.J., Linhares, D.C.L., 2018. Porcine reproductive and respiratory syndrome monitoring in breeding herds using processing fluids. Journal of Swine Health and Production 26, 5.
Prickett, J., Simer, R., Christopher-Hennings, J., Yoon, K.J., Evans, R.B., Zimmerman, J.J., 2008a. Detection of Porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: a longitudinal study under experimental conditions. Journal of veterinary diagnostic investigation 20, 156-163.
Prickett, J.R., Kim, W., Simer, R., Yoon, K.-J., Zimmerman, J., 2008b. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. Journal of Swine Health and Production 16, 86-91.
Rossow, K.D., 1998. Porcine reproductive and respiratory syndrome. Vet Pathol 35, 1-20.
Rossow, K.D., Bautista, E.M., Goyal, S.M., Molitor, T.W., Murtaugh, M.P., Morrison, R.B., Benfield, D.A., Collins, J.E., 1994. Experimental porcine reproductive and respiratory syndrome virus infection in 1-, 4-, and 10-week-old pigs. Journal of veterinary diagnostic investigation 6, 3-12.
Rossow, K.D., Shivers, J.L., Yeske, P.E., Polson, D.D., Rowland, R.R., Lawson, S.R., Murtaugh, M.P., Nelson, E.A., Collins, J.E., 1999. Porcine reproductive and respiratory syndrome virus infection in neonatal pigs characterized by marked neurovirulence. Vet Rec 144, 444-448.
Silva, G., Schwartz, M., Morrison, R., Linhares, D.C., 2018. Monitoring breeding herd production data to detect disease outbreaks. In, Monitoring and Surveillance 2.0 seminar. In: 49th Annual Meeting of the American Association of Swine Veterinarians., San Diego, Calif., 9-10.
Silva, G.S., Schwartz, M., Morrison, R.B., Linhares, D.C.L., 2017. Monitoring breeding herd production data to detect PRRSV outbreaks. Prev Vet Med 148, 89-93.
Xiao, Z., Batista, L., Dee, S., Halbur, P., Murtaugh, M.P., 2004. The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. Journal of virology 78, 5923-5933.
Yeske-Livermore, L., O’Neil, K., Main, R., Zimmerman, J., 2014. Improved pre-weaning surveillance using oral fluids: pilot study. 44th AASV meeting, Dallas, Texas, 317-318.
You May Also Like


