Family oral fluids: understanding when and how much to pool
Pooling is highly beneficial if goal is to test more families within a fixed monitoring budget.
April 22, 2021

Introduction
Monitoring remains a key component of PRRS control programs 1. The use of aggregate sample types for ascertaining PRRS status of herds has made monitoring cheaper, easier, less laborious, and more sensitive, especially in low prevalence scenarios 2,3. Aggregate sample types such as processing fluids and oral fluids are currently major sample types submitted to the veterinary diagnostic laboratories from breeding herds for PRRS monitoring 4.
Family oral fluids (FOFs), an aggregate sample type, are oral fluids obtained from a dam and her piglets. Usually gotten around the time of weaning, this sample type has been demonstrated to be practical 5 and useful in determining the PRRS status of due-to-wean piglets (Almeida, 2019).
For herds that have capacity for weaning more than one room on a weekly basis, testing individual FOF samples can have significant cost implications. This is especially true in low prevalence situations where only a handful of crates are likely to give a positive PRRS PCR test result. The strategy of pooling aggregate sample types to save cost while maintaining a comfortable degree of sensitivity on testing has been demonstrated on processing fluid samples 2,7.
The primary objective of this study was to investigate the effect of pooling FOFs at different levels on the probability of PRRSV detection by RT-rtPCR (Reverse transcriptase, real time polymerase chain reaction).
Methodology
PRRSV-positive FOF samples were obtained from PRRSV-Unstable breeding herds. PRRSV-negative FOF samples were obtained from PRRSV-negative herds. The statuses of these samples were confirmed before proceeding with the experiment.
30 PRRSV-positive samples were conveniently selected and tested at different pooling levels: undiluted, 1:3, 1:5, 1:10, and 1:20 (1:3 means 1-part positive PRRSV sample pooled with 2 parts negative PRRSV sample). These pooling levels were repeated 6 times (replicates) for each sample and tested for PRRSV RNA by RT-rtPCR.
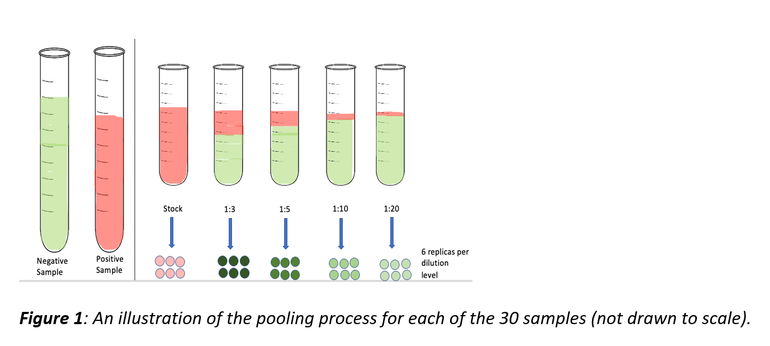
The probability of detection was the outcome of interest and was defined as the fraction of the 6 replicates that was positive on PCR for each dilution level for each sample.
For the purposes of analysis, the samples were further grouped into 3 categories based on PCR Cycle thresholds (Ct) values. Simply put, a Ct value is the number of PCR cycles it took for fluorescence signals to cross a threshold; they are a good proxy of the quantity of PRRSV RNA within a sample. The lower the Ct value, the higher the quantity of PRRSV RNA in the sample tested. According to the specifications for the PCR kit used for this experiment, when a Ct value is £ 40, it is considered PCR-positive, otherwise, it is negative.
Results
Five samples were excluded from the analysis because the PCR results of the undiluted replicates of these samples were above 40. The 25 samples left were grouped into one of 3 categories based on their Ct values. The subcategories were A. Ct < 34 (n = 8) B. 34 ≤ Ct ≤ 36 (n = 8) C. Ct > 36 (n = 9).
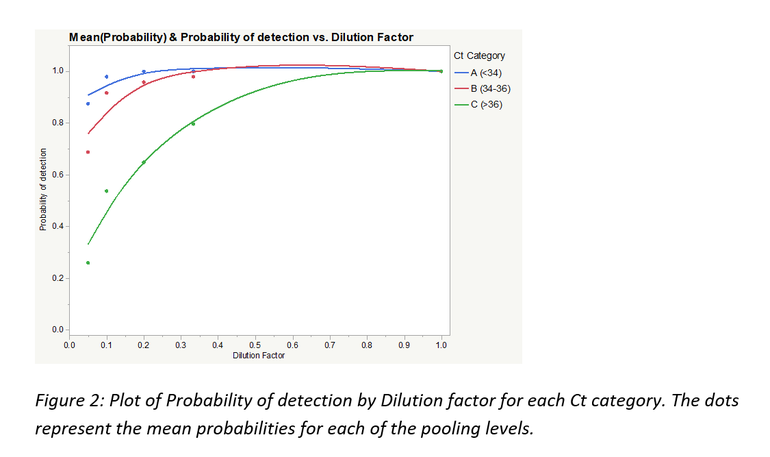
Figure 2 is a plot of the probability of detection by the dilution factors 1, 0.33, 0.2, 0.1 and 0.05 (representing undiluted, 1:3, 1:5, 1:10 and 1:20) for each Ct category.
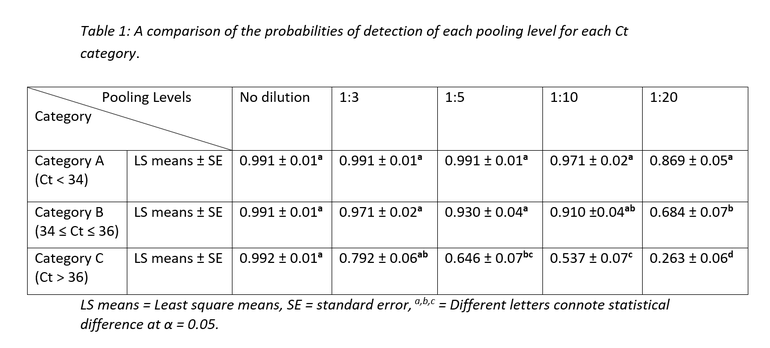
Table 1 demonstrates the probability of detection for each of the pooling levels for each of the Ct categories.
Discussion
The primary aim of this study was to demonstrate the effect of pooling FOFs on the probability of PRRSV RNA detection by PCR. The approach taken was a quite conservative one as samples from PRRS-negative herds were used for the pooling exercise as opposed to PRRSV-negative samples from the same herds from which the positive FOF samples were sourced. The tables and figures shown above represent the most extreme dilution situations where there is only one positive sample in any given pool.
As seen from table 1 and figure 2, Ct category, and by extension, viral concentration per sample significantly influenced the probabilities of PRRSV RNA detection. For Ct category A, there was no statistically significant difference in probability of detection across all pooling categories even though some numerical differences in probability of detection can be observed between pooling levels. Ct category C had the poorest numbers of the 3 categories with the probability of detection dropping to below 30% at 1:20 pooling level.
To the best of our knowledge, there has not been a published study relating Cts of FOFs to possible predictors. Time since outbreak has been demonstrated to be associated with Ct values of processing fluids 8. Logical guesses could also be made using recent laboratory data from the herd.
From the data we currently have for breeding herds sampled (unpublished), FOF Cts are more than 95% of the time either in categories A or B.
As would often be the case, there could be more than one positive sample within and/or between pools; this significantly improves the probability of detection. A simulation was performed to demonstrate this for each of the Ct categories using laboratory data from this study. Figures 3 to 5 represent the results of those simulations.
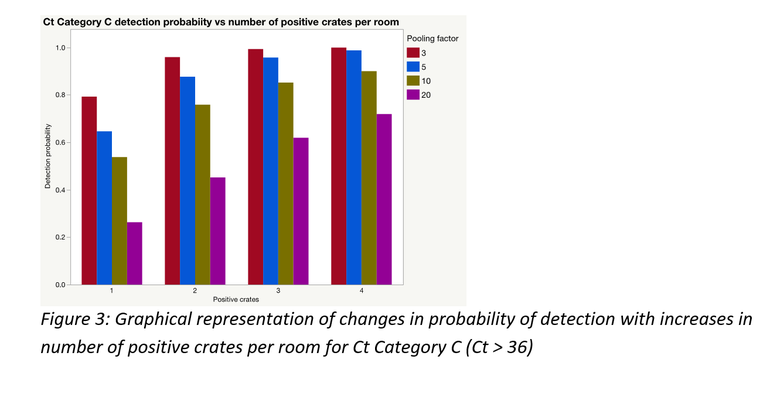
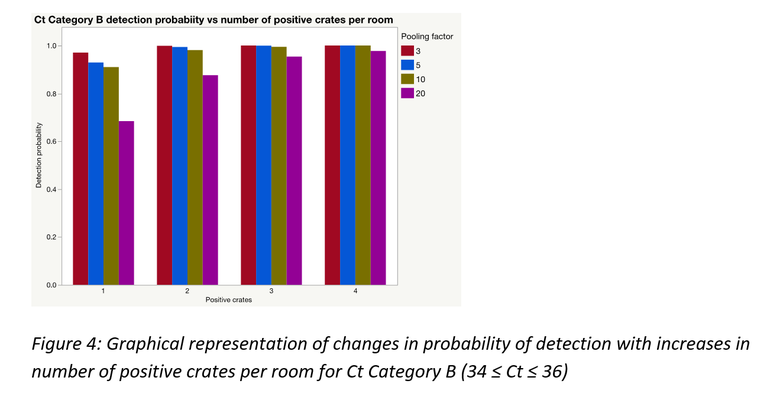
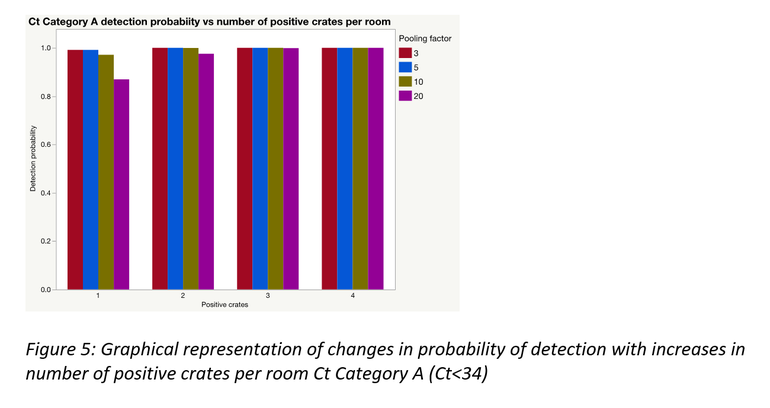
Pooling is highly beneficial if the goal is to test more families within a fixed monitoring budget. Mathematically, under certain assumptions, only a portion of a farrowing room could be sampled and pooled at an assumed between crate prevalence, to be at least 95% certain of obtaining a PCR positive result for PRRSV, if present.
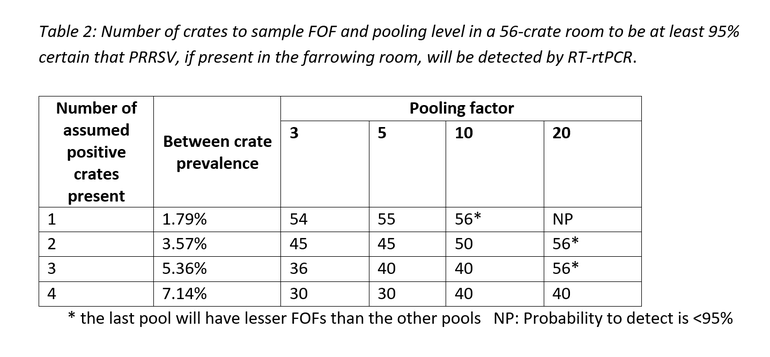
As an example, below is a tabular breakdown of the number of crates to sample and pool in a 56-crate farrowing room to be at least 95% certain of obtaining a PCR positive result for PRRSV, if present.
Conclusion
The importance of consistent monitoring to ascertain herd PRRS status cannot be overemphasized. Pooling FOFs is a valid cost-effective option for PRRSV monitoring because more families in a barn can be tested on a budget (especially helpful in very low prevalence scenarios) without sacrificing significant sensitivity. A decision to pool and the level of pooling is reliant on the level of sensitivity farm management is comfortable with.
Conflict of Interest
The authors of this work declare no conflict of interest.
Acknowledgement
The authors wish to thank the Iowa Livestock Health Advisory Council (ILHAC) for sponsoring this project.
Sources: Onyekachukwu Osemeke, Marcelo Almeida, Giovani Trevisan, Gustavo Silva, Arka Ghosh, Daniel Linhares, Iowa State University, who are solely responsible for the information provided, and wholly own the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
References
1. Martín-Valls GE, Hidalgo M, Cano E, Mateu E. Testing of umbilical cords by real time PCR is suitable for assessing vertical transmission of porcine reproductive and respiratory syndrome virus under field conditions. Vet J. 2018;234:27-29. doi:10.1016/j.tvjl.2018.01.008.
2. López WA, Zimmerman JJ, Gauger PC, et al. Practical aspects of PRRSV RNA detection in processing fluids collected in commercial swine farms. Prev Vet Med. 2020;180:105021. doi:10.1016/j.prevetmed.2020.105021.
3. Nunes de Almeida M. Improved porcine reproductive and respiratory syndrome virus surveillance in swine breeding herds. Grad Theses Diss. January 2020. doi:10.31274/etd-20210114-104.
4. Trevisan G, Linhares LCM, Crim B, et al. Macroepidemiological aspects of porcine reproductive and respiratory syndrome virus detection by major United States veterinary diagnostic laboratories over time, age group, and specimen. Shaman J, ed. PLoS One. 2019;14(10):e0223544. doi:10.1371/journal.pone.0223544.
5. Almeida MN, Rotto H, Schneider P, et al. Collecting oral fluid samples from due-to-wean litters. Prev Vet Med. 2020;174. doi:10.1016/j.prevetmed.2019.104810.
6. Almeida M. Title: PRRSv Detection over Time in Different Age Groups in Breeding Herds Attempting to Produce PRRSv-Negative Piglets at Weaning: Part 2: Suckling Pig Population (NPB 18-161).
7. Vilalta C, Baker J, Sanhueza J, et al. Effect of litter aggregation and pooling on detection of porcine reproductive and respiratory virus in piglet processing fluids. J Vet Diagnostic Investig. 2019;31(4):625-628. doi:10.1177/1040638719852999.
8. Trevisan G, Jablonski E, Angulo J, Lopez WA, Linhares DCL. Use of processing fluid samples for longitudinal monitoring of PRRS virus in herds undergoing virus elimination. Porc Heal Manag. 2019;5(1):1-5. doi:10.1186/s40813-019-0125-x.
You May Also Like



