This information has the potential to improve the welfare of animals as well as the economic cost associated with current PCV2 management strategies.
December 18, 2018

By Lianna R. Walker, University of Nebraska-Lincoln PhD student
Unraveling the host genetic factors involved in disease resistance or rather disease tolerance is not only daunting, but deemed by some as a nearly impossible task. Despite being a trait of moderate heritability, it is exceptionally complex, polygenic and highly susceptible to environmental influences, making circumstances of natural infection difficult to replicate experimentally.
This problem is exceptionally relevant when evaluating the elusive pathogen responsible for a set of symptoms with prominent economic impacts, such as post-weaning multisystemic wasting syndrome. This pathogen is porcine circovirus 2, the smallest known virus capable of infecting mammals.
Although PCV2 is the causative agent of this group of well-known porcine circovirus associated diseases, not all infected pigs will develop clinical disease. In fact, majority of the pigs remain sub-clinically infected, allowing for continued global spread and infection of pig populations worldwide. PCVAD incidence and severity has been illustrated by anecdotal field data to vary between breeds and individuals, suggesting host genetics may be an important factor in PCV2 pathogenesis.
To investigate the role of host genetics in PCV2 susceptibility, a large-scale experimental infection was conducted with about 1,000 maternal crossbred pigs infected with a PCV2b strain. These pigs represented 14 genetic lines generated by seven genetic programs. The challenge extended for 28 days post infection, with weights and blood collected weekly. The amount of virus in the blood (viremia), PCV2-specific antibody level (IgM and IgG), and average daily gain (kilograms per day) were measured weekly throughout the challenge. From the weekly viremia measurements, the total amount of virus during the challenge (viral load) was also calculated. Each animal was genotyped with the Porcine SNP60 BeadArray consisting of about 60,000 DNA markers.
A genome-wide association analysis using these 60,000 genotypes was performed for each of the measured traits (viremia, viral load, antibody response and ADG). The proportion of phenotypic variation accounted for by the DNA markers varied between traits and time points post-infection. The largest proportion was for viral load, with host genotype accounting for 64% of the phenotypic variation. Two major genomic regions located on host chromosomes 7 and 12 were found to account for 17.7% of this variation, representing putative Quantitative Trait Loci for PCV2 viral load.
The QTL located on chromosome 7 is adjacent to a group of genes with major impact on the host immune response known as the Swine Leukocyte Antigen Class II complex. The QTL on chromosome 12 was originally on an unplaced chromosomal fragment in the previous genome reference assembly (Sscrofa 10.2). Linkage disequilibrium estimates predicted the location of the top marker (ALGA0110477) to be on the proximal end of chromosome 12. With the release of the new swine reference assembly (Sscrofa 11.1), this region is no longer missing, and the placement of the QTL has been confirmed with ALGA0110477 explaining 6.2% of the phenotypic variation in viral load.
Since the region detected on chromosome 12 was not present in the previous assembly, it had not been annotated or investigated in swine. A 20 Mb chromosome segment generated from long-read DNA sequencing that provided precise location of ALGA0110477 was utilized to dissect the chromosome 12 QTL region. A combination of gene prediction methods, RNA-sequencing and extensive genomic sequencing uncovered five positional candidate genes located within 150 kb of ALGA0110477 and over 60 novel polymorphisms within or upstream of these genes. Association analyses including the genotypes of about 600 DNA markers mapped to the chromosome 12 segment and these 60 novel polymorphisms, highlighted a polymorphism (SYNGR2 p.Arg63Cys) that alone accounted for more than 20% of the phenotypic variation in viral load. Additionally, SYNGR2 p.Arg63Cys had a significant impact on measures of weekly viremia and ADG, with the favorable genotype (Cys63/Cys63) associated with decreased viremia (Figure 1) and increased ADG (Figure 2) following PCV2 challenge.
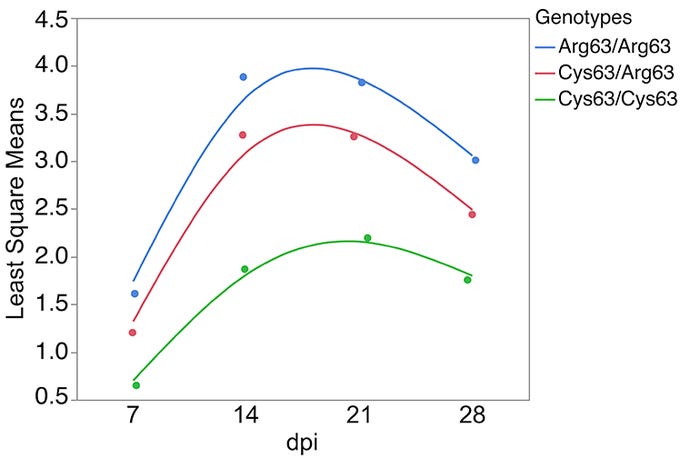
Figure 1: Least square means of SYNGR2 p.Arg63Cys genotypes across measures of weekly viremia following PCV2 infection (n=268)
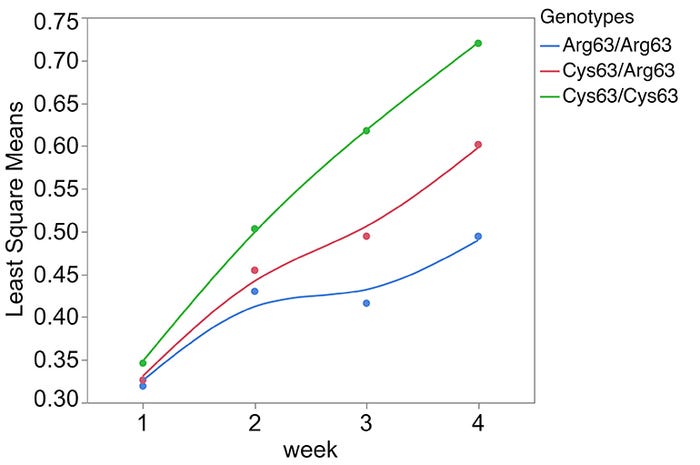
Figure 2: Least square means of SYNGR2 p.Arg63Cys genotypes across measures of weekly average daily gain (kilograms per day) following PCV2 infection (n=268)
The function of Synaptogyrin-2 (SYNGR2) is not well characterized, but it is hypothesized to function in membrane trafficking and transport and is expressed in nearly every cell of the body. Research in rats identified two crucial domains of the SYNGR2 protein required for proper function. The SYNGR2 p.Arg63Cys polymorphism is located within one of these critical domains and results in a non-conservative amino acid substitution from arginine to Cysteine. This domain is fairly conserved across mammalian species and the Cys amino acid at this location is unique to swine.
Specifically, the Cys allele was present in a single haplotype in the resource population. Based on phylogenetic analysis, this haplotype is closest to those found in Duroc and Pietrain. Since the Cys allele is predominant in these paternal breeds where selection emphasis is placed on growth, there may have been unintentional selection for PCV2 tolerance due to subtle variation in growth caused by early less virulent PCV2 strains.
An in vitro validation model using the porcine kidney 15 cell line was employed to provide direct experimental evidence of the role of SYNGR2 in facilitating PCV2 multiplication. First, SYNGR2 expression was decreased by more than 80% in PK15 cells and then these cells were infected with PCV2b. A significant reduction in viral replication was observed in the cells with reduced SYNGR2 expression compared to control PK15 cells. Additionally, CRISPR-Cas9 was utilized to generate an edited PK15 clone (E1) homozygous for a 106bp deletion in the SYNGR2 gene, predicted to result in a non-functional SYNGR2 protein. Subsequent infection of the edited E1 PK15 cells with PCV2b once again resulted in a significant reduction in PCV2 titer, with almost no evidence of viral replication following infection (Figure 3).
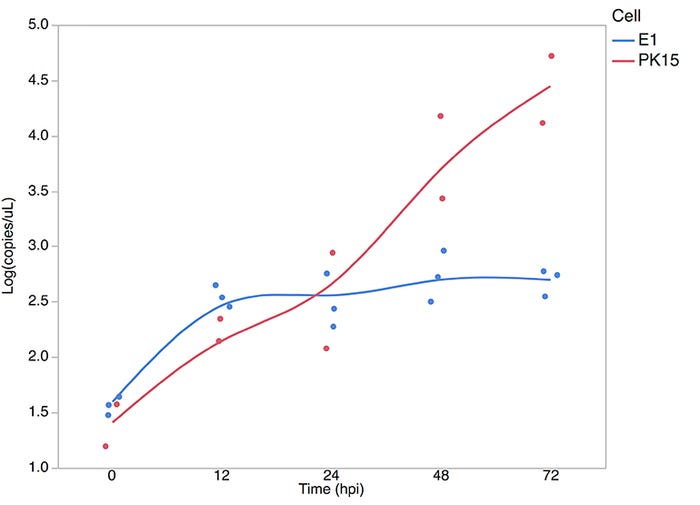
Figure 3: PCV2 titer expressed as log transformed viral copies/uL present in the supernatant across hours post infection of edited PK15 (E1) and wild-type PK15 cells
All together, these findings provide direct experimental evidence of the role of SYNGR2 in influencing PCV2 multiplication and strongly implicates SYNGR2 p.Arg63Cys as a putative functional polymorphism that controls PCV2 susceptibility. This information has the potential to improve the welfare of animals as well as the economic cost associated with current PCV2 management strategies by providing the ability to assess PCV2 susceptibility prior to exposure or development of PCVAD. Since this is a natural DNA variant, animals with elevated tolerance to PCV2 can be identified and selected for early on, potentially representing a new and cost-effective method of PCVAD prevention.
This study was led by scientists from the Departments of Animal Science and Statistics of the University of Nebraska-Lincoln and involved researchers from the U.S. Meat Animal Research Center, Dalhousie University and University of Alberta. The project was supported by Agriculture and Food Research Initiative Competitive Grant No. 2016-09374 from the USDA National Institute of Food and Agriculture, by a grant from Genome Canada and by a Strategic Investment Program Grant from the University of Nebraska-Lincoln. The results of the study were recently published in PLOS Genetics, https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007750.
You May Also Like



