S. suis are diverse bacteria and the diversity of S. suis isolates demonstrated by serotyping and MLST testing must be understood in greater detail in regard to pathogenicity.
April 9, 2019

By April A. Estrada, Connie Gebhart, Stephanie Rossow and Douglas G. Marthaler, University of Minnesota; and Marcelo Gottschalk, University of Montreal, Canada
Streptococcus suis has recently re-emerged as a significant cause of increased mortality in piglets and growing pigs. Non-disease-causing commensal S. suis strains normally reside in the upper respiratory tracts of pigs while pathogenic strains cause meningitis, arthritis, endocarditis, polyserositis and septicemia. S. suis can also act as an opportunistic pathogen, causing respiratory disease in the presence of other bacterial or viral pathogens. Due to differences in the pathogenicity of S. suis strains, diagnosing and selecting the correct strain for vaccine development can be difficult. In addition, multiple pathogenic types exist, and vaccination with one pathogenic strain may not lead to cross-protection for other pathogenic strains.
Subtyping methods for S. suis
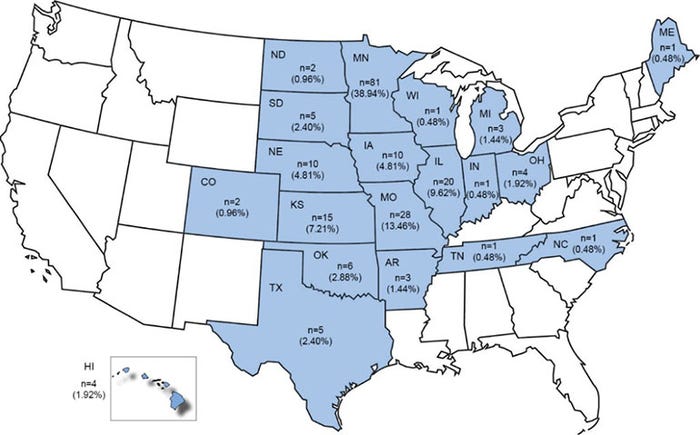
Typing methods may potentially be used to differentiate between S. suis pathogenic and commensal strains. Serotyping and multilocus sequence typing are two common methods for subtyping S. suis strains. Traditionally, serotyping is a serologic test, but polymerase chain reaction-based tests have been developed. There are 35 known S. suis serotypes (1-34, and 1/2), with serotypes 2 and 3 reported to be recovered from diseased pigs in North America (1, 2). MLST is a molecular subtyping approach and 1,170 different S. suis sequence type (ST) profiles have been identified to date. In North America, ST25 and ST28 were reported to be commonly recovered from diseased animals (3). Although the highly pathogenic ST1 strain is more prevalent in Europe (4), ST1 strains are also a concern in North America. However, information on the serotype and ST diversity of other S. suis strains found in the U.S. is limited. Our latest research has improved our understanding of this diversity (Figure 1).
Pathotype classification of S. suis
Since pigs naturally carry S. suis on their tonsils, multiple isolates can be recovered from a single healthy or diseased pig, making it difficult to identify the disease-causing strains. In our study, we classified S. suis isolates as pathogenic, possibly opportunistic or commensal based on clinical history and specimen type. Pathogenic isolates were obtained from neurologic and systemic tissues submitted to the University of Minnesota and Kansas State Veterinary Diagnostic Laboratories and were reported by pathologists as the primary cause of disease. Possibly opportunistic isolates were obtained from lung samples while commensal isolates were recovered from healthy pigs, with no known history or current control methods for S. suis disease in the farm.
Characterization of pathogenic isolates
We characterized 208 S. suis isolates obtained between 2014 and 2017 from diseased and healthy pigs in the U.S. by serotyping and MLST. Serotyping identified 19 different serotypes with 14 serotypes recovered from pigs with neurologic or systemic disease. The predominant pathogenic serotypes identified in diseased pigs were 1/2, 2, and 7, all of which have been previously associated with S. suis disease (2, 5). Our results illustrated that serotype 1/2 is more common in our U.S. isolate set, and 80.4% of serotype 1/2 isolates were linked to clinical disease. Further odds ratio analysis by pathotype classification identified serotypes that could be differentiated as pathogenic, possibly opportunistic or commensal pathotypes (Table 1).
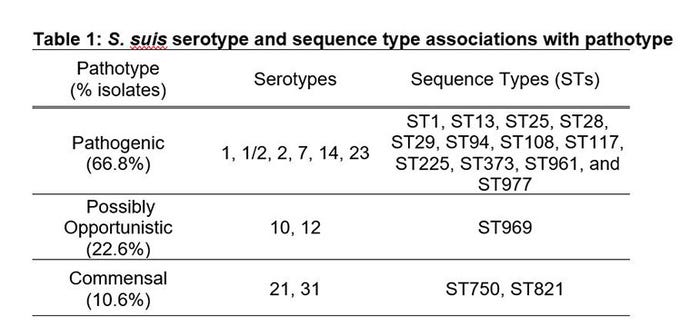
MLST identified 58 different STs with 24 STs recovered from pigs with clinical disease. The predominant pathogenic STs identified in diseased pigs were ST1, ST28, ST94 and ST108. All ST1 isolates and 80.8% of the ST28 isolates were classified as pathogenic. Further odds ratio analysis by pathotype classification identified STs that could be differentiated as pathogenic, possibly opportunistic or commensal pathotypes (Table 1).
S. suis are diverse bacteria and the diversity of S. suis isolates demonstrated by serotyping and MLST testing must be understood in greater detail in regard to pathogenicity. Typing of S. suis strains indicated ST as a stronger predictor of pathotype compared to serotype. This research to identify pathogenic strains can improve the prevention and control of S. suis by selecting strains for development of diagnostic tests (e.g. virulence-associated gene profiling) and S. suis vaccines. Typing of strains can also provide general information on the prevalent subtypes affecting herds and can potentially be used to monitor changes in serotype and ST distributions.
References
1. Messier S, Lacouture S, Gottschalk M. 2008. Distribution of Streptococcus suis capsular types from 2001 to 2007. Can Vet J Rev Veterinaire Can 49:461–462.
2. Fittipaldi N, Fuller TE, Teel JF, Wilson TL, Wolfram TJ, Lowery DE, Gottschalk M. 2009. Serotype distribution and production of muramidase-released protein, extracellular factor and suilysin by field strains of Streptococcus suis isolated in the United States. Vet Microbiol 139:310–317.
3. Fittipaldi N, Xu J, Lacouture S, Tharavichitkul P, Osaki M, Sekizaki T, Takamatsu D, Gottschalk M. 2011. Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg Infect Dis 17:2239–2244.
4. Schultsz C, Jansen E, Keijzers W, Rothkamp A, Duim B, Wagenaar JA, van der Ende A. 2012. Differences in the Population Structure of Invasive Streptococcus suis Strains Isolated from Pigs and from Humans in the Netherlands. PLoS ONE 7:e33854.
5. Gottschalk M, Lacouture S, Bonifait L, Roy D, Fittipaldi N, Grenier D. 2013. Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Québec, Canada. Vet Microbiol 162:819–825.
Source: University of Minnesota and University of Montreal, who is solely responsible for the information provided, and wholly owns the information. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like



