January 5, 2016

Influenza continues to be one of the most costly and under-recognized diseases in pigs. Part of the challenge in controlling influenza is the limited information available on influenza transmission at the farm level and what factors contribute to the introduction, maintenance and dissemination of influenza virus.
A University of Minnesota study evaluated the role that the different pig subpopulations play at supporting influenza infections. Researchers monitored replacement animals that had been recently introduced (less than four weeks), replacement animals that had been in the gilt development unit for a prolonged period (more than four weeks) and piglets for 12 months in five breeding herds.
Five commercial pig-breeding herds located in the Midwestern U.S. were selected for this study. Selection criteria included breed-to-wean herds with: a) confirmed influenza A virus infection by real-time reverse transcriptase polymerase chain reaction (RRT-PCR) within the past year, b) presence of an on-site gilt development unit, and c) introduction of external replacement animals into a defined isolation area.
Each farm was visited monthly for 12 months, and the overall sampling period spanned from November 2011 to December 2012. At each visit, three pig subpopulations were sampled: a) replacement females, resident on-farm for less than four weeks (new gilts), b) replacement females, resident on-farm for more than four weeks (gilts), and c) neonatal pigs less than 21 days of age (piglets). Adult animals (sows) were not included since prior studies consistently reported a low probability of influenza detection in this subpopulation. Due to schedules for delivery of replacement females, eligible populations of new gilts were only present at 21 of the 60 sampling events. A diagram of the study design can be seen in Figure 1. Information on management and control practices for IAV was recorded during the last visit.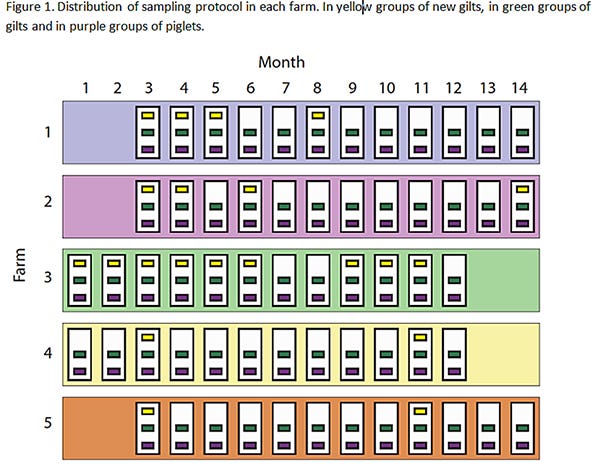
Five swine-breeding herds (largest rectangles, 1 to 5) in the Midwest were sampled between November 2011 (month 1) and December 2012 (month 14). White rectangles represent sampling visits (12 per farm), and the smallest rectangles (n = 141) indicate the groups of pigs sampled. Groups are colored based on pig subpopulation: yellow (new gilts, n = 21), green (gilts, n = 60), and purple (piglets, n = 60). Missing group-rectangles indicate that there were no new gilts on that visit. Groups were assumed nested within sampling events, and sampling events were assumed nested within farms.
Thirty pigs were selected from each pig subpopulation (new gilts, gilts and piglets) and sampled individually using a nasal swab. Sample size (n = 30) was estimated based on a 95% confidence to detect at least one positive sample if prevalence was 10% or higher at the subpopulation level.
Nasal swabs were first screened for IAV by RRT-PCR on pools of three samples. If a pool tested positive, the samples comprising the pool were tested individually. All samples in a negative pool were considered negative. Viral RNA was eluted using 50 microliter of each sample into 50 microliter elution buffer. Primers targeting the matrix (M) gene and a reagent kit were used to detect IAV. PCR mix containing 5 microliter RNA, 12.5 microliter 2X buffer, 1.0 microliter 25X enzyme mix, 1.67 microliter detection enhancer, 5 picomol of each primer and 1.5 picomol of probe was run at 45 degrees Celsius for 10 minutes, followed by 95 C for 10 minutes, and 45 cycles at 94 C for one second and 60 C for 30 seconds. Fluorescence was recorded at 60 C, and a sample was considered positive if the cycle threshold was lower than 40. Positive samples with a CT value of 35 or lower were used for IAV isolation, and each IAV isolate was subtyped based on the hemagglutinin and neuraminidase combination.
Data collected in the survey was summarized by farm, and three independent variables were taken into consideration for statistical analysis: a) pig subpopulation, b) farm and c) annual quarter (first to fourth). The association between IAV detection and subpopulation was analyzed at the group level where a group was defined as the subpopulation of pigs that was sampled during a given visit and considered positive if one or more swabs within the group tested positive to IAV.
Logistic regression models
Using gilts as the reference group, an unconditional logistic regression model was used to measure the crude association between the outcome and pig subpopulations, farm and annual quarter. Finally, fixed and mixed logistic regression models were compared to estimate the associations between IAV infection and subpopulation adjusting by farm, annual quarter and sampling event. Subpopulation and annual quarter were included as fixed effects in the models, and clustering variables (e.g., farm and farm visit) were included as random effects. Given the study design (Figure 1), researchers assumed that samples clustered by sampling visit, farm and annual quarter. It was also assumed that pig subpopulations were nested within sampling visits, and that sampling visits were nested within farm.
There were 4,190 nasal swabs collected from 141 groups of pigs. At least one influenza-positive nasal swab was found in 19.9% (n = 28) of the sampled groups, and 7.7% (n = 324) of all nasal swabs tested positive. After adjusting by annual quarter and sampling event, the odds of testing influenza positive were 7.9 (95% CI 1.4, 43.9) and 4.4 (95% CI 1.1, 17.1) times higher in groups of new gilts and piglets compared to groups of gilts, respectively. Results indicate that new gilts and piglets had higher odds of testing IAV-positive than gilts in swine breeding herds and that season influences IAV infection in pigs.
Piglets tested positive at least once in all farms; new gilts only tested positive in Farm 1 and Farm 3; and all gilts tested negative in Farm 5 (Figure 2). One hundred and twenty-four IAVs isolates were recovered and 123 of them were successfully subtyped. Subtypes H1N1, H1N2 and H3N2 were identified, and more than one IAV subtype was isolated in all farms over time.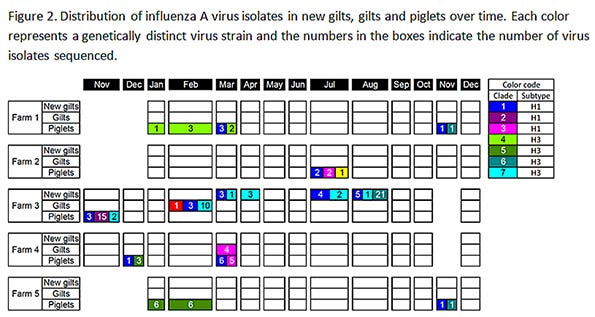
Furthermore, research results also show the co-circulation of multiple influenza viruses in a given farm or subpopulation over time. Up to three genetically distinct viruses were identified in each farm at a given time, which highlights the challenges to effectively control influenza.
Based on these findings, researchers recommend that IAV control strategies be aimed at preventing infection before gilts are introduced into the farm and in pigs prior to weaning.
You May Also Like



