Conventional sampling methods are based on individual sampling, which is time consuming, expensive, requires expertise and is not effective to detect disease at low prevalence.
November 6, 2017

By Will Lopez, Giovani Trevisan, Jeff Zimmerman, Karen Harmon, Phil Gauger, Luis Gimenez-Lirola, Christopher Rademacher and Daniel Linhares, Iowa State University; and Jose Angulo, Zoetis USA.
Disease monitoring and surveillance systems are based on sampling a subset of the population to estimate disease frequency (or to detect disease) at the herd level. Effectiveness of monitoring and surveillance systems can be measured using specific metrics including:
• Time to detect outbreak: can be improved by increasing the frequency of sampling (e.g. daily, weekly or monthly).
• Limit of detection at the population level: can be improved by increasing the sample size (e.g. 30 samples detect disease at ≥ 10% prevalence).
• Accuracy of detection: depends on herd sensitivity and specificity of tests being used.
Therefore, optimum monitoring scheme is fast, practical, affordable and effective. Conventional sampling methods are based on individual sampling (e.g. individual blood samples), which is time consuming, expensive, requires expertise and is not effective to detect disease at low prevalence.
The objective of this column is to describe the use of processing fluids-based sampling to monitor breeding herds for porcine reproductive and respiratory syndrome virus. For the purpose of this column, we define processing fluids as a composite sample formed of tissue exudate derived from the process of piglet castration and tail docking (Figure 1).
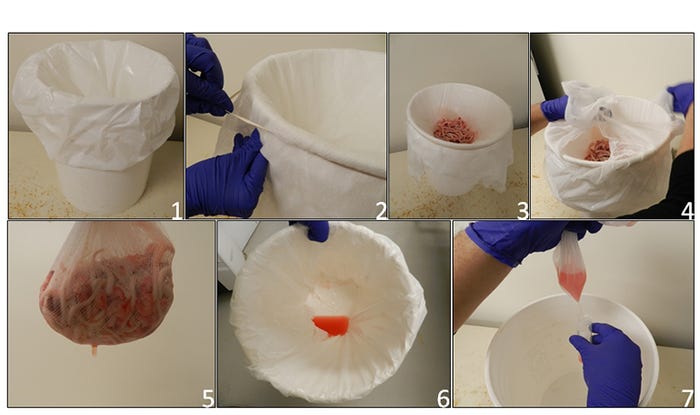
Figure 1: Guidelines to obtain processing fluids from neonates:1. Plastic bag in clean bucket; 2. Cheese cloth placed and fixed over mouth of bucket to hold tissues and allow fluid to pass through to plastic bag; 3. Collection of tissues from the processing of the litters; 4-5. Tissues are removed after collection; 6. Processing fluid recovered in plastic bag; 7. Fluid transferred from plastic bag into a sterile tube for transportation.
In order to describe the use of processing fluids-based sampling to monitor breeding herds for PRRS virus, we compared monitoring programs using this sample methodology against monitoring programs using the conventional sampling method of 30 serum samples, or 30 tail blood swabs at processing time. To do so, the samples collected by either method were taken from the same pig populations on the same days.
In our field studies, we collected processing fluid samples from commercial breed-to-wean (sow) herds that were doing active surveillance for PRRS virus and we compared each processing fluid sample to, either the 30 individual blood serum samples or the 30 individual tail blood swabs, depending on the preferred method by the veterinarian of each herd. Those individual samples were collected from the same pig population on the same day of processing fluid collection. That is what we call “matching set of samples.” We also tested the processing fluids for anti-PRRS antibodies by ELISA (IDEXX PRRS X3 ELISA) and also submitted the samples for ORF-5 sequencing. All the samples were tested at the Iowa State University Veterinary Diagnostic Laboratory.
Results
The preliminary field work done by our group, which is available in the 2017 Iowa State University James McKean Swine Disease Conference proceedings1,2, has documented the ability to detect PRRSV RNA by PCR in processing fluids with a higher herd sensitivity (greater capability to detect PRRSV-positive pigs) than matching 30 individual blood sera, or 30 tail blood swabs when these sample types were pooled in groups of five. Processing fluids had an overall frequency of detection for PRRSV of 66% (21 positives out of 32 samples) whereas the corresponding individual samples (blood sera and tail blood swabs) pooled in groups of five had a lower frequency of detection for the virus of 31% (10 positives out of 32 sets of samples). The frequency of detection of PRRSV for the blood sera and tail blood swabs in terms of total pools was of 7% (13 positive pools out of 192 total pools tested). In all sampling attempts, the collection of processing fluids had a 100% of success rate and the task was easily conducted by farm staff. The volume of processing fluids retrieved in every attempt was enough to perform multiple diagnostic tests.
Moreover, we have been able to obtain PRRSV sequences from 15 out of 18 samples (83%), which supports the hypothesis that PRRSV RNA is relatively stable and in high quantities in processing fluids obtained under field conditions, from farms undergoing virus elimination.
Furthermore, we have also been able to detect anti-PRRSV antibodies by ELISA, and PCV2 DNA by PCR in processing fluids, which suggests that this sample type may be useful for other diagnostic investigations/applications.
We recommend keeping processing fluids refrigerated or frozen shortly after collection, following similar guidelines available for oral fluids. Preliminary work at ISU has demonstrated no loss in PCR sensitivity when samples are stored for up to five days at 4 degrees Celsius (refrigerator temperature), and/or after one freeze/thaw cycle.
Implications
Preliminary results of ongoing field studies support the conclusion that obtaining processing fluids from up to 700 piglets per sample, is easy, practical and economical. This allows monitoring PRRSV and other infectious diseases as frequently as daily, which significantly improves monitoring and surveillance systems for infectious diseases at a relative low cost.
The increased frequency of sampling that can be achieved with processing fluids and the considerable increase in the number of pigs (can test them all!!) that can be tested in one single sample is a major improvement in developing monitoring and surveillance systems for infectious agents capable of disease detection at very low prevalence.
Processing fluids-based sampling is, therefore, a simple method to screen herds for PRRSV in a practical, fast and effective fashion. Farms undergoing virus elimination can use processing fluids to understand when PRRSV-negative pigs are being produced. Evidence of continued PRRSV circulation (positive processing fluids) will alert farm managers and staff to switch to more aggressive bio management practices to stop virus circulation and avoid further dissemination within the herd. Likewise, consistently negative processing fluids inform producers and veterinarians that additional surveillance is necessary, e.g., to start monitoring due-to-wean piglets with family oral fluids (subsequent column in NHF Daily will be available on this method) or other methods, to fully ensure that weaned piglets are truly negative before being shipped from sow farms.
References
1. W. A. Lopez et al. Processing Fluids for Detection of PRRS Activity in Neonates. 2017 ISU James D. McKean Swine Disease Conference. P. 65
2. W. A. Lopez et al. Processing Fluids, Blood Serum, and Tail Blood Swabs to Detect PRRSV RNA and PCV2 DNA by PCR-Based 2017 ISU James D. McKean Swine Disease Conference. P. 69
You May Also Like



