A resistant bacterium is the one that is not affected by an antimicrobial. However, behind that definition are two different approaches that are worth knowing.
May 9, 2017

By Carles Vilalta DVM, PhD, University of Minnesota
Occasionally, and with more frequency, we see and hear about deaths related to multi drug resistant bugs, commonly called superbugs by the media. This scenario and its implications (Figure 1) has increased society’s awareness about the use of antimicrobials, both in humans and animals, and their potential role in selecting resistant bacteria. The higher the use of antimicrobials, the higher the risk of selecting resistant bugs.
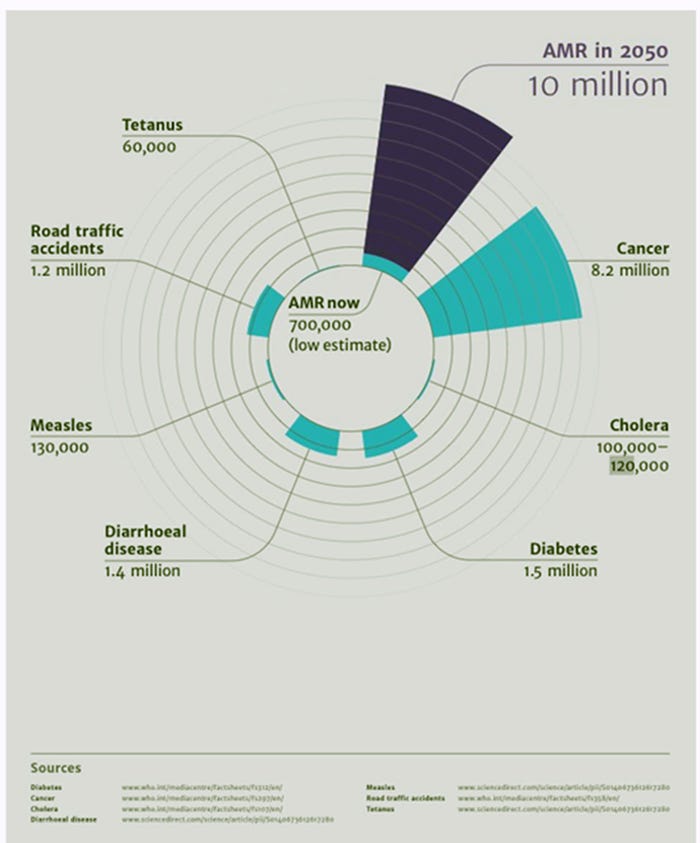
Figure 1: Deaths attributable to antimicrobial resistance compare with other causes of death every year currently (light blue) and predicted (purple). Source: Forbes
But do we know what it means when we hear the term resistant in either a farm context, or in lab results? Actually, the basic definition is very straight forward. A resistant bacterium is the one that is not affected by an antimicrobial. However, behind that definition are two different approaches that are worth knowing.
The first way of defining bacteria as resistant is from the pharmacological approach. You might recognize that this approach was used when the result from the lab or the data you are looking at has the categories of Susceptible (S), Intermediate (I) or Resistant (R) or a given minimum inhibitory concentration (MIC). The MIC is the minimum amount of drug needed to inhibit the growth of bacteria on a Petri dish or in a tube. It will be expressed as milligrams per milliliter or micrograms per microliter. The higher the MIC, the more resistant the bacteria can be. That means when treating a sick animal we would need more antimicrobial to reach the site where the bacteria are. Sometimes that difficulty can be overcome by increasing the dose, treating more frequently, or prolonging the treatment.
That would work until we reach the point where the dose would be too high, causing the liver and kidneys of the animal to be unable to get rid of more of the compound. The drug then starts to accumulate in the body, intoxicating or eventually killing the animal. To help veterinarians choose the right treatment some organizations divided the MIC distribution into three breakpoints based on the likelihood of success of the treatment, or according to achievable concentrations of that specific antimicrobial in the site of action. Thus, S, I and R mean high, medium, or low likelihood of success in the treatment of that antimicrobial in the registered dose against that specific pathogen. To facilitate the diagnostic job, some labs use what is called infusion disks, disks embedded with a certain concentration of drug and placed onto a culture of the specific microbe of interest. With those plates we can test different drugs and infer their likelihood of treatment success, using the radius of the inhibition zones. Finally, the breakpoints are specific to the particular drug and microbe combinations tested, and should not be used to infer breakpoints for other combinations.
The second method is from the bacterial point of view (microbiological approach) and uses a breakpoint or cut-off value that separates the MIC distribution of the bacteria that do not express certain resistant genes (wild type) versus the distribution of the microbes that express those mechanisms. Basically, here we will call those bacteria resistant those have resistant genes, creating the potential to be antimicrobially resistant. To establish that separation we need to categorize the bacteria according to the resistant genes that they may possess and link that to the MIC of each specific strain against that specific compound. An important complication in determining gene-based resistance is the potential for the transfer of a resistance gene between bacteria or bacterial families (Figure 2). These transfers make it harder to use an antimicrobial on any bacteria in possession of those resistant genes. Acquisition of resistance against a certain type of drug can cause the bacteria to also be resistant to another compound. This is what is called cross-resistance.
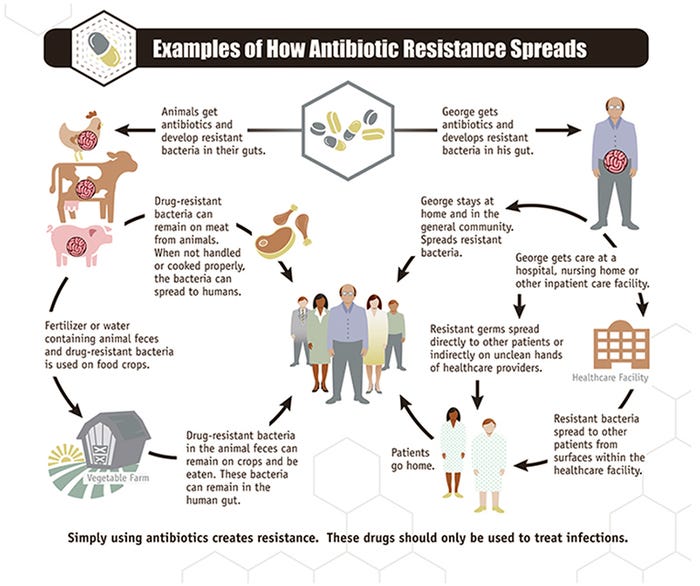
Figure 2: Pathways of resistant genes spread. Source: CDC. Centers for Disease Control and Prevention.
Even though the term “resistant” is understood as a microbe being resistant against an antimicrobial, we can see that how the label was reached affects its meaning. In some cases, bacteria have the genetic potential to avoid the chemical compound and in some other cases not enough drug will reach the infection site to reduce the bacteria to a level that the immune system can take care of. The more information we have about the underlying cause of resistance, the better. Mistreatments and under treatments can shift bacterial selection toward a resistant strain of the bacteria.
If you are further interested in this topic you might be interested in the short video made by the Harvard Medical School, “The Evolution of Bacteria on a ‘Mega-Plate’ Petri Dish” that shows bacteria overcoming a 1,000-fold specific killing concentration.
You May Also Like



