April 2, 2012
Diagnostic tools and their applications for porcine reproductive and respiratory syndrome (PRRS) virus continue to evolve. PRRS virus sequencing is increasingly being utilized as a tool to enhance pork producers’ understanding of PRRS virus that is actively circulating within a herd, production system or geographic region.
Sequencing is commonly conducted following the detection of PRRS virus in instances where knowing the strain(s) of the circulating virus is useful. Sequencing data can help swine veterinarians and producers understand the source, evolution and movement of PRRS virus into, within and between herds.
Figure 1 illustrates the number of PRRS virus positive samples being sequenced at the Iowa State University Veterinary Diagnostic Laboratory and the time from the initial case submission to sequencing results being reported. As you can see, there has been a significant increase (> 200%) in the amount of sequencing being conducted and a considerable decrease (≈50%) in the time to complete these sequencing analyses.
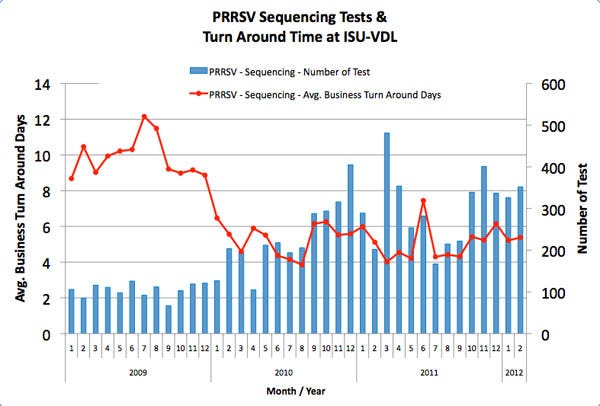
This increase in PRRS virus genetic sequencing analyses being conducted is due to a surge in individual producers utilizing this tool, as well as sequencing data being used in the number of area regional control projects initiated over the past two years. The improved turnaround time has been driven by efforts to streamline these otherwise more complex diagnostic procedures at the laboratories.
Historically, serum from viremic pigs has been the sample type of choice for herd-level PRRS virus detection and genetic sequencing. More recently, oral fluid diagnostics have been increasing as an effective tool for detecting PRRS virus. However, determination of the PRRS virus sequence from oral fluid samples has been problematic. In analyzing more than 3,000 diagnostic records from PRRS virus positive serum and oral fluid samples, investigators found sequencing success to be 84% and 50% in serum and oral fluid samples, respectively.
Paul Thomas, third year veterinary student at Iowa State University, and others recently conducted a pilot study evaluating the effects of various sample collection and handling interventions on sequencing success in oral fluids. This work found that sequencing success was significantly affected by the temperature in which samples were kept upon collection through transport to the laboratory. Sequencing success was 96%, 86% and 73% for samples being frozen, refrigerated or held at room temperature, respectively.
These data suggest that PRRS virus sequencing success rates in oral fluids can be improved by freezing samples immediately upon collection and maintaining this cold chain through submission to the diagnostic laboratory.
Although there is still much to learn, the recent advancements in PRRS virus oral fluid diagnostics suggest that oral fluids may become the sample type of choice for herd-level PRRS virus diagnostic and monitoring applications in the future. These advancements include the recent introduction of the PRRS virus oral fluid ELISA assay, improvements in PRRS virus PCR methods, and we are beginning to understand the opportunities to improve PRRS virus sequencing success rates using oral fluids.
Contact Rodger Main, DVM at [email protected]
You May Also Like


