A lot of terminology gets thrown around when one starts discussing genetic engineering technologies. Kristin Whitworth and Randall Prather offer a quick overview.
July 5, 2016
The frequency at which new technologies are being developed and introduced to both the scientific community and the public is astonishing. This is especially true of genetic engineering technologies.
First, there was enu mutagenesis, then transgenes, conditional transgenes, homologous recombination, embryonic stem cells, knockouts, knockins, and now gene editing. Terms like selectable markers, meganucleases, zinc-finger nucleases, TALENs, CRISPRs and base pairs, previously confined to scientific journals, are now showing up in popular press articles on a routine basis. Here we would like to provide a quick review of what some of these terms mean and how they relate to one of the newer technologies that is being discussed — gene editing.
To begin the discussion of gene editing, we must first answer the question what is a gene? In a general sense, a gene is a segment of DNA that codes for the production of RNA. This RNA then is translated into protein. So, DNA makes RNA makes protein. This process is referred to as “Central Dogma” in scientific literature. Simply put, if you change the DNA (adding, subtracting and substituting), then a different RNA is made, and a different protein can be made, or no protein is made at all.
As simple as ATC
The most common building blocks or letters of DNA are As, Ts, Cs and Gs (they are also called bases). In the pig, there are probably about 3 billion bases in the genome. The bases are arranged in sequential order and are packaged to create a chromosome. Pigs have 19 pairs of chromosomes (humans have 23 pairs). Each member of a pair of chromosomes is derived from different parents. That means that for each gene, there are two copies; one copy from the mother and another copy from the father. Each of these copies is referred to as an allele. One allele is inherited from the father through the sperm, and the other allele is inherited from the mother through the egg. If the DNA from a single cell is stretched out in a single strand, it would be about six feet long. The ability to package these strands into chromosomes and then fit them into a nucleus of a cell is pretty remarkable considering that a normal cell is on the order of 0.01 millimeters in diameter.
A gene is composed of a section of a strand of DNA that, depending on the sequence of the bases (As, Ts, Cs and Gs), has many different parts. The first part that we will discuss here is the promoter. The promoter can be thought of as a rheostat for a light. It can be off, or it can be on. Further, the “on” can be slow, medium or fast. In the case of a gene, this means that there can be no RNA production (and hence no protein), or it can mean that there is a small, medium or large amount of RNA made (and generally, but not always, a small, medium or large amount of protein that is made).
The next part of the gene are the exons. Exons contain the sequence of bases that code for the amino acids. A triplet code is used to direct the sequence of amino acids in a protein. This means that the sequence of three bases codes for a single amino acid. The next three bases code for the next amino acid. There is a special triplet usually in the first exon beyond the promoter that codes for the amino acid methionine and is what is called the “start”, because this is generally the first amino acid in the string of amino acids that comprise a protein. The end of the string of amino acids is coded for by another specific triplet code called the “stop”.
Each gene can have multiple exons separated by introns (intervening sequences). When the RNA is made from the DNA, these introns are spliced out so that the exons are joined together. Genes vary in length (number of bases) and the number of exons and thus the resulting proteins have different sizes, structures and functions.
Protein factory
We now need to talk about the importance of the sequence of amino acids. One analogy that might work (remember all analogies break down) is to compare a cell to a self-replicating factory. Given the correct inputs (energy, iron, plastic, etc.), the factory can replicate itself. The machines in the factory that are used to make a new factory are made by the factory. Each of the parts of the machines in the factory are a specific string of amino acids that is called a protein. Long strings of amino acids make big parts (proteins), and short strings make smaller parts (proteins). Since each amino acid has a slightly different three-dimensional structure, stringing different sequences of amino acids together will make a different three-dimensional shape (a part) when the string of amino acids fold together.
These three-dimensional parts need to fit together correctly so they function correctly as a tool. An example might be a hammer. The synthesis of the handle part might be directed from one gene, while the head of the hammer is directed from another gene. The handle and the head then must both fit together and have two functional parts (the handle that is the correct shape to function and the head that is the correct shape to function). Some parts may have multiple functions. The head of the hammer may have an end for hammering nails and another end that is used to pull nails. For the most part, each factory (cell of the body) has a base set of directions (blueprints or DNA), parts and tools, or machines that are made for the cell to replicate. Since each cell type in the body is a little different, a different set of genes (blueprints) is turned on in each cell type so that a different set of parts/tools/machines are made. These different sets of parts determine what the cell does (Figure 1).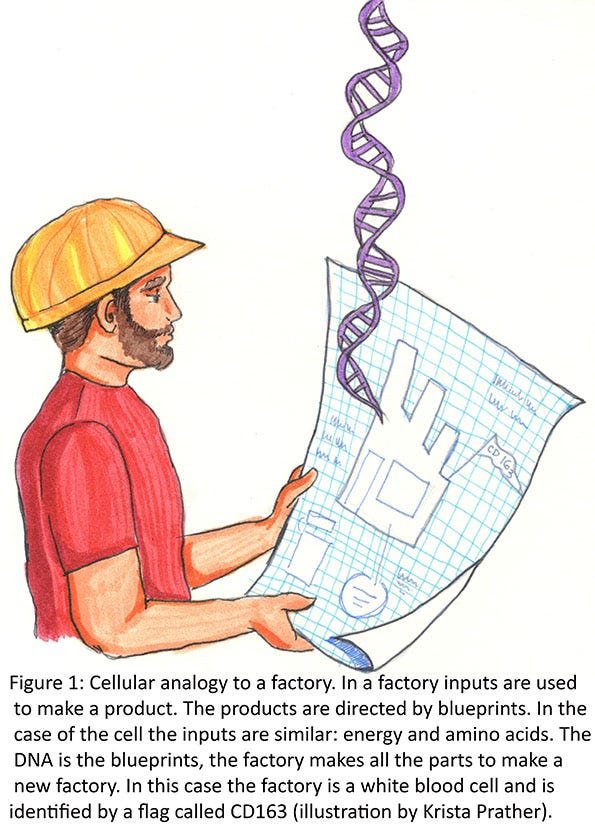
In summary, the sequence of bases in the promoter determines, in a cell-specific manner, if a gene is “turned on”, the degree to which it is “turned on”, and where RNA synthesis starts. RNA is made and then introns are spliced out, thus joining the exons. The spliced RNA then is translated into a specific sequence of amino acids which makes up the protein.
Raising the PRRSV flag
Now we’ll give an example that has relevance to porcine reproductive and respiratory virus. The PRRSV binds to a protein on the surface of alveolar macrophages. The alveolar macrophage is a white blood cell in the air sacs of the lungs. The alveolar macrophage has a specific set of genes that are turned on that code for a specific set of proteins that make the cell into an alveolar macrophage. One specific gene that is uniquely turned on in the alveolar macrophage is a gene called CD163 (cluster of differentiation 163). The CD163 gene codes for a unique sequence of amino acids. This unique sequence of amino acids folds into a unique three-dimensional structure, and in this case, is put on the outside of the factory like a flag, i.e. it sticks out of the factory (or outside the cell).
The PRRSV uses the unique three-dimensional shape of that flag created by the unique sequence of amino acids to infect the cell. You may ask “Why don’t other cell types get infected by PRRSV?” The answer is because the other cell types, e.g. skin epithelial cells, don’t turn on CD163 and they don’t have the flag on the outside of the cell. Without the CD163 flag, the PRRSV doesn’t recognize the cell and thus can’t infect the skin cell.
To create the pigs that are resistant to PRRSV, we used gene editing technology. The most common gene editors are zinc-finger nucleases, Tal-effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/Cas9). All three systems contain at least two elements. The first is a method to target a unique sequence in the genome. In our case, this was a region of an exon in the CD163 gene. In the case of the CRISPR/Cas9, we used a guide of 20 bases (letters) to target a region in exon 7 that was unique to CD163, i.e. this sequence was not found anywhere else in the pig genome. The second element is a nuclease. As the name implies, a nuclease cuts nucleotide bonds (these are the bonds that hold the individual bases [letters of the genome] together). The nuclease we used was CRISPR-associated 9 (Cas9). The guide targets the nuclease to a specific region of the genome and cuts it.
Generally, there are DNA repair mechanisms that immediately act to repair the cut in the DNA. It is thought that usually the DNA is correctly repaired, but the guide and nuclease are still present, and so it cuts it again. This process is repeated until a mistake is made in the repair. Examples can include adding a base or removing a base. Once the target is cut and repaired with a mistake, the guide no longer recognizes this specific region of the gene and does not try to cut it again. In our case we had one pig that had seven bases added to one allele, and another 11 bases removed from the other allele. Remember, the genome of the pig is some 3 billion bases. In this particular pig, we changed 18 bases. Since neither of these gene edits is divisible by three, this results in what is called a “frame-shift” edit, because the triplet code for adding the amino acids is shifted and a non-sense sequence of amino acids follows until a “stop” is encountered. The RNA then is degraded or translated into a short protein. If translation occurs, the protein often becomes misfolded and is degraded (Figure 2).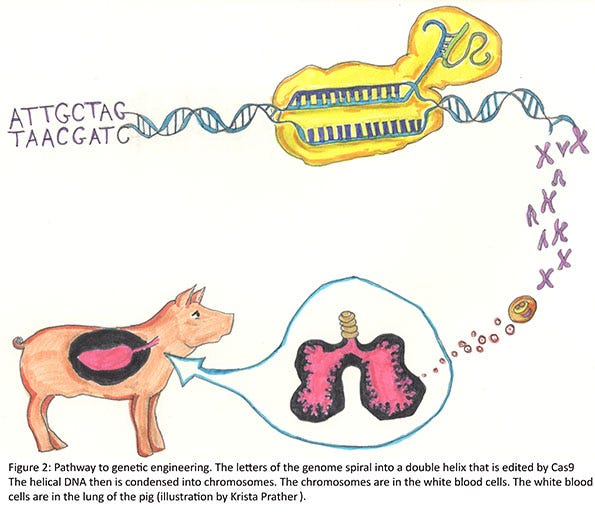
Knockout punch
The big advantages of the gene editors is that they are highly efficient at cutting targeted DNA. In addition, the CRISPR/Cas9 system is quick and easy to use. So now you can imagine changing a few bases of the genome. If this change is at the start of a gene, you can knockout a gene, i.e. make it so no protein is produced. If the edit is strategically placed in the middle of a gene, you might be able to remove a middle portion of a gene, and thus the middle amino acids of a protein. When the gene editor is used in combination with donor DNA, the donor DNA might be inserted to make a very specific change in the bases, via something called homologous recombination (that is a topic for another day).
The gene editing technology discussed above is distinct from transgenesis. Transgenic organisms, by definition, contain a gene from another species. The addition of a transgene generally means that all the components of a gene are introduced into the host genome. This includes a DNA sequence that is composed of a promoter, exons and introns. Genes can be thousands of bases (letters) long. In contrast, the gene edits that were described above can be as little as a single base that was changed to knockout a gene.
One of the questions that we are often asked is: “Isn’t a knockout of a gene always bad?” True, if some genes are knocked out, a detrimental or lethal phenotype will result. But the answer to that question is, “No, not all knockouts are detrimental or lethal.” In fact, when the technology to knockout genes became available for mice, phenotyping centers were created because often there was either no phenotype or an unexpected phenotype. Within mice, there is redundancy such that not every gene is necessary for a normal life. We have knocked out a number of genes in the pig with little effect on the phenotype (examples include: CD163, sialoadhesin, alpha-1, 3-galactysyltransferase, and cytidine monophosphate-N-acetylneuraminic acid hydroxylase-like protein: the latter two are to create pigs that can serve as organ donors for humans). There are some genes in which a knockout is detrimental to development or even lethal, but in the case of CD163, the pigs appear quite normal.
To go back to the factory analogy; the sequence of bases in the DNA amounts to the blueprints for the factory to replicate itself. Genetic engineering is similar to redesigning a factory to perform unique tasks. To do the new task you need to add or remove parts. To add or remove parts you need to change the blueprints, or in this case, the sequence of bases (letters) in the strands of DNA.
We are living in an exciting era of huge technological and scientific advances. The ability to selectively edit genes in domestic animals is amazing. To use those genetic edits to improve the health, well-being and productivity of pigs is a natural outgrowth. We hope this lesson in DNA Editing 101 helps readers to understand the genetic editing technology so they can put it in context and determine the extent of its use.
You May Also Like



