Genomic Selection Shows Promise in Swine Breeding
Genetic improvement has played a role in improving nearly every production efficiency trait evaluated in livestock, including pigs. The rapid evolution of gene technology allows swine breeders and commercial pork producers to make breeding decisions based on gene marker technology
April 15, 2010
Genetic improvement has played a role in improving nearly every production efficiency trait evaluated in livestock, including pigs. The rapid evolution of gene technology allows swine breeders and commercial pork producers to make breeding decisions based on gene marker technology, once thought to be applicable only to researchers leading the “genome revolution.”
Pig breeders have been using gene marker technology since the early 1990s to remove deleterious genes like the halothane gene (HAL), which causes porcine stress syndrome, and the napole gene (RN-) from their herds.
Several new gene marker tools are now commercially available at relatively low costs for traits including number of pigs born, feed efficiency, growth rate, backfat depth and pork quality. These tools are but the first wave of the genomic technology that will continue to revolutionize pork production efficiency.
Evolution of Quantitative Tools
For many years, pork producers relied on the physical attributes and growth rate measures (phenotypic evaluation) to evaluate the sires of their next pig crop. Later, quantitative genetics and selection indexes were employed to more accurately estimate an animal's genetic merit by using a number of economically important traits and a limited number of relative's records.
In the '50s and '60s, testing stations sprang up to standardize the environment in which boars' performance was measured and indexed. This information helped guide producers toward sires that would improve the productivity of their herds. Development of the backfat probe improved measurement accuracy and, hence, lean percentage.
Soon, whole-herd testing helped identify family lines that consistently excelled in economically important production traits. A new statistical procedure called best linear unbiased prediction (BLUP), used in mixed models for the prediction of random effects, allowed breeders to take the performance information from numerous individuals and combine it with the genetic relationship between the animals. This accounting for the relationships between animals was a major advancement in estimating genetic merit.
During the 1970s and 1980s, advancements in ultrasonic technology provided measurement of a variety of traits, such as backfat, loin muscle area and depth and intra-muscular fat to help predict lean percentage and meat quality traits.
Commercial producers can now conduct a genetic evaluation of their sow herd to identify females with the most desirable estimated breeding values (EBVs) to produce replacement gilts. Breeders commonly use more complex computer tools, such as mate selection programs, to optimize genetic progress and track inbreeding accumulations.
Marker-Assisted Selection
Breeding value computations have become even more complex with the advent of marker-assisted selection (MAS). By combining molecular genetic information and traditional quantitative procedures, the rate of genetic improvement has taken another step forward.
Screening animals for marker genotypes is relatively easy with blood or tissue samples (whole blood, blood blotter cards, ear notches, docked tails or tissue obtained through an approved ear tagging system) as sources of DNA. These screening tests are relatively inexpensive and as technology improves, more tests will be available and prices typically decline.
Currently, commercial markers are available to improve several performance traits. The MC4R, HMGA1 and CCKAR (cholecystokinin type A receptor) markers will improve growth rate, feed efficiency and carcass lean percentage.
Specifically, the MC4R locus impacts growth and leanness in the pig. There are two alleles or variants of this marker — A and G. The A allele is associated with fast growth, while the G variant is associated with lean and efficient growth.
Pigs that are homozygous (i.e. AA) for the fast-growth alleles have been shown to reach market weight three days sooner than pigs that are homozygous for the lean allele (GG). Pigs selected for the MC4R lean alleles (GG) will have 8% less backfat and eat significantly less feed per pound of gain.
The HMGA1 gene marker is highly associated with backfat and lean growth, and the CCKAR is associated with feed intake, hunger control and obesity.
Molecular tests are also available to help develop and improve breeds and genetic lines for meat quality using the PRKAG3 and CAST markers.
The PRKAG3 marker, observed in all major pig breeds, is associated with muscle glycogen content and meat quality. The CAST marker — an abbreviation for Calpastatin — is responsible for inhibiting enzymes called proteases, which affect meat tenderness.
Reproductive Trait Focus
Increasing litter size is one way to improve production efficiency — requiring fewer sows and less feed per pig produced. Selection for increased litter size is responsible for the large gains breeders have made in this trait in the last 20 years. Two new markers — the ESR (estrogen receptor) gene and the EPOR (erythopoietin receptor) gene — are now commercially available.

The ESR gene was first discovered in Meishan pigs, a notable impact of three Chinese breeds imported into the United States in the early 1980s. The ESR gene marker is associated with litter size and has been used for many years by a large pig breeding company. This gene marker is effective in the Large White or Yorkshire breeds and crossbred females of those two breeds.
The EPOR marker gene is associated with uterine capacity and litter size.
To learn more about these markers and the associated genetic tests, go to www.geneseek.com (Geneseek, Lincoln, NE) or www.dnalandmarks.ca (DNA Landmarks, Quebec, Canada).
Tapping Genomic Technology
Producers should capitalize on this genomic technology by testing herd boars and stud boars for these genetic markers and using them in pure-line matings. The backfat, growth rate and meat quality status of all boars used to develop terminal sire lines should also be known. Similarly, the gene marker status for litter size should be known for all boars used to make maternal purebred matings.
With marker test results, breeders can determine the frequency of the alleles, good and bad, for the breeds or lines of sires providing semen. This information can also help determine whether further testing of females from a breed or line is necessary.
In addition, breeders can determine which alleles they would like to “fix” to be sure that all animals in a breed or line have two copies of the preferred allele. In this manner, preferred animals can be selected and those without the desirable alleles can be culled.

60,000 Gene Markers
Some genetic suppliers are already using from 300 to 3,000 markers from their private research. Therefore, genomic selection may be available in the next 5-10 years, even for the medium to large breeders.
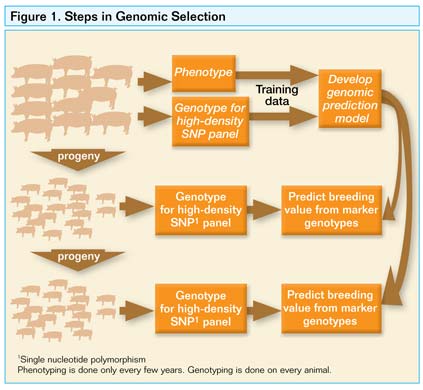
Genomic selection is a means to simultaneously select on many genetic markers across the genome that together explain most of the genetic differences between animals. As a result, selection of the best animals can be based on markers without measuring phenotype on each animal or before phenotype is even measured (Figure 1).
To enable genomic selection, researchers have now designed a single nucleotide polymorphism (SNP) chip which contains 60,000 genetic markers across the pig genome. This was accomplished by successful efforts to obtain the first draft genome sequence in the pig.
The genome sequence is the first information on all genes in the pig and represents a powerful tool for future use. Outcomes from the sequencing and the SNP chip are likely 5-10 years away, but producers should begin using the genetic tests now and plan to employ the other technology in the future.
Precision Phenotyping
Which is better — 30 pigs/sow/year (p/s/y) or a sow that raises 26 p/s/y through five or six parities?
Which is better — 10% faster growth or 10% faster growth with 20% less feed?
Do we want pork that tastes better, has less fat and perhaps more vitamins and nutrients?
Precision pig production will require us to identify new traits more accurately. Genomic selection for traits such as feed intake, feed efficiency, days-to-first service, rebreeding interval and sow longevity offer real opportunities to turn pig production into precision production.
Breeding stock suppliers will employ complicated selection systems (Figure 2) to manage data from all ends of the production pyramid. Coordinated initiatives will allow them to refine genetic programs for commercial production, to maximize carcass value and, in turn, help food companies deliver a variety of meat products that will find favor with consumers.
The tools are available to accelerate genetic improvement, including statistical advancements, phenotyping and genomic technology. The benefactors will be pork consumers.
You May Also Like



