Antibiotics have powerful effects on the composition and structure of the pig’s gut microbiome. These changes are likely linked to changes in the health of both the intestine locally and the host systemically.
April 24, 2018

By James Lowe, University of Illinois College of Veterinary Medicine i-Learning Center director
The benefit of antibiotics to animal and human health is both profound and unquestioned.
From our historical “pathogen-centric” view of disease, where specific microbes caused specific diseases, antibiotics were the unquestioned king of managing infectious disease. As technology has improved, so has our understanding of how microbes interact with both humans and animals, which has led to a more sophisticated view of infectious disease.
We have learned that microbes (bacteria, viruses and protozoa), commonly referred to as the microbiota, along with macroparasites (worms), have extensive communication with each other and with the host they reside on and in. This “cross talk” is beneficial to the host, as it helps control the pathogens both by direct challenge (microbe-microbe) and by enhancing the host local immune response.
In this “host-centric” view of disease, it is a lack of balance in the microbe-host interaction that results in disease. Pathogens can only succeed in the host-microbiota ecosystem when the stability of the ecosystem is disrupted from stress, diet changes, the addition of new microbes or other environmental factors. Exposure to the pathogen alone is not enough to cause disease; it takes a failure of the host-microbiota ecosystem to maintain homeostasis.
Gut health is a good example of this complex interaction. The normal development of the intestine relies on complex interactions with the microbiota that colonizes in the intestine. This exposure begins at birth and continues throughout the lifetime of the individual.
While less is known in pigs, the process has been studied extensively in humans. Babies born via caesarean section have very different gut microbiomes than do vaginally delivered babies.1,2 Information remains limited regarding the source of these microbes.
We investigated whether specific strains of bifidobacteria in the maternal intestinal flora are transmitted to the infant’s intestine. Fecal samples were collected from 17 healthy mother-and-infant pairs (vaginal delivery: 12; C-section delivery: 5). This “colonizing” microbiome has long-term consequences for health in humans, including asthma3 and Type 1 diabetes.4,5 In addition, extended antibiotic treatment in the perinatal period results in a higher risk of late-onset Group B Streptococcus sepsis, necrotizing enterocolitis and overall mortality in the neonatal period.4,6
While the impact of antibiotics during the neonatal period is significant in humans, less is known about pigs. Our group has attempted to understand the factors that drive the development of the microbiome of the pig’s intestine though a series of experiments.
In our first experiment, we employed a cross-fostering strategy to determine which sources of microbiota were the most important in establishing the piglet’s gut microbiome, and if different microbiomes resulted in different levels of “gut health” in piglets. Piglets were given high-quality colostrum from their birth dam or a foster dam upon birth. Twenty-four piglets from two litters (12 pigs per litter) were randomly assigned to one of three treatment groups according to the source of colostrum and postcolostral milk feeding for the subsequent 21 days.
Treatment 1 (T1; n=8) received colostrum and postcolostral milk feeding from their own dam. Treatment 2 (T2; n=8) were litter exchanged at birth to receive colostrum from a foster dam for 24 hours and then returned to their own dam for postcolostral milk feeding the subsequent days. Treatment 3 (T3; n=8) were litter exchanged at birth to receive colostrum and postcolostral milk feeding from a foster dam, and they remained with the foster dam for the subsequent days.
Each piglet was allowed to suck colostrum for equivalent times. The piglets were observed to exhibit vigorous teat sucking and subsequent satiation. All sows were clinically healthy with no history of receiving any antibiotics prior to farrowing. None of the piglets was administered antibiotics during the experimental period. All piglets were weighed directly after birth and before being euthanized at Day 21.
At farrowing, a single colostrum sample, fecal and vaginal swab were collected from each sow. Fecal swabs were collected from each piglet on Day 0 and Day 21. At Day 21, a group of 12 piglets was humanely euthanized. Samples of the luminal contents, tissue and mucosa were collected along the gastrointestinal tract. To determine the microbiome of each sample, high-throughput sequencing was used on the Illumina MiSeq platform. To determine the function of the gut wall, quantitative real-time polymerase chain reaction analysis was also performed to quantify the expression of toll-like receptors 2, TLR 4, TLR 10, tumor necrosis factor alpha, interferon gamma, and interleukin 4 and IL 10.
We found that microbial communities varied according to the GI biogeographical location, with colon being the most diverse section. Bacterial communities in both maternal colostrum and vaginal samples were significantly associated with those present in the fecal samples of piglets, suggesting that, like in humans, the mother is an important source of the initial colonizing microbiome in pigs.
Cross-fostering did not affect bacterial communities present in the piglet GI tract. The mRNA expression of TLR and inflammatory cytokines changed (P<0.05) with biogeographical location in the GI tract. Higher mRNA expression of TLR and inflammatory cytokines were observed in ileum and ileum-associated lymph tissues.
This study suggests an impact of colostrum and maternal microbial communities on the microbiota development and mucosal immune gene expression in the newly born piglet that closely mimics the process that has been described in humans. So, what about the impact of antibiotics in neonatal piglets?
In a second experiment, we investigated the impact of five different antibiotics given at birth on growth, microbiome development and the prevalence of antimicrobial resistance genes in suckling pigs. We used a randomized complete block design. Forty-eight litters were blocked to one of six treatments (N=8) by farrowing day, dam parity group and litters with a minimum of nine pigs. Within the litter, all pigs received the same treatment. Pigs were weighed, and treatments were administered within 24 hours of age, after litters were balanced for size.
Treatments were as follows: control (saline 1 cc), tulathromycin (2.5 mg/kg IM), ceftiofur crystalline free acid (5 mg/kg IM), ceftiofur hydrochloride (5 mg/kg IM), oxytetracycline (22 mg/kg IM) and procaine penicillin G (33,000 units/kg IM). Two pigs per litter were individually identified, and weights and deep fecal swabs were collected at days 0 (pretreatment), 5, 10, 15 and 20 (see Figure 1).
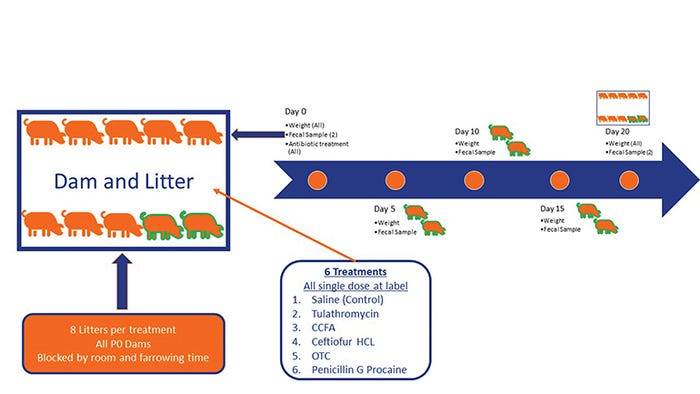
Figure 1: Experimental design to determine the impact of giving antibiotics to piglets at birth on their growth, their fecal microbiome composition and the development of antibiotic resistance.
As in the first study, samples were subjected to high-throughput sequencing used on the Illumina MiSeq platform to determine the microbiome and the prevalence of 7 ARG in each sample. Antimicrobial treatment had no effect on individual weight gain, litter weight gain or mortality. Unlike the data from prolonged use in neonatal humans, antibiotics had no effect on the fecal microbiome composition or diversity during the neonatal period (see Figure 2).
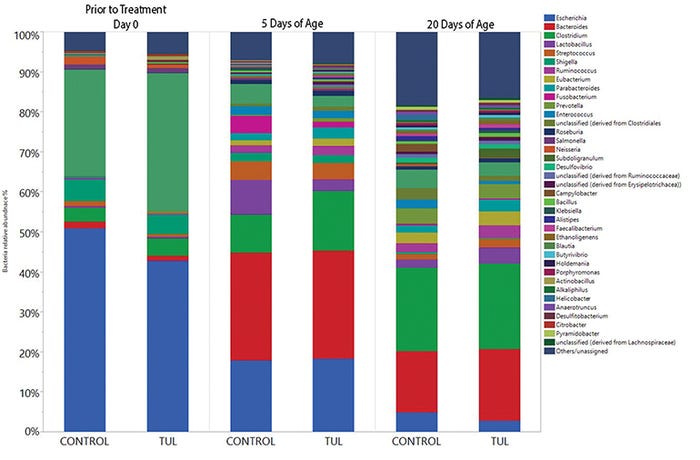
Figure 2: Antibiotic impact on fecal microbiome composition 20 days after administration in neonatal pigs. Pigs were sampled at birth and then treated with tulathromycin or saline (control). Pigs had fecal samples collected on days 5 and 20. Colors represent different bacterial genus. There were no significant differences between treatments, but the composition of the fecal microbiome changed over time.
Interestingly, there were also no major increases in ARG with antibiotic admiration at birth. Only one antibiotic, TUL, resulted in any increase in ARG during the entire neonatal period (see Figure 3). Of the seven ARG investigated, only four were elevated in samples from the TUL-treated pigs.
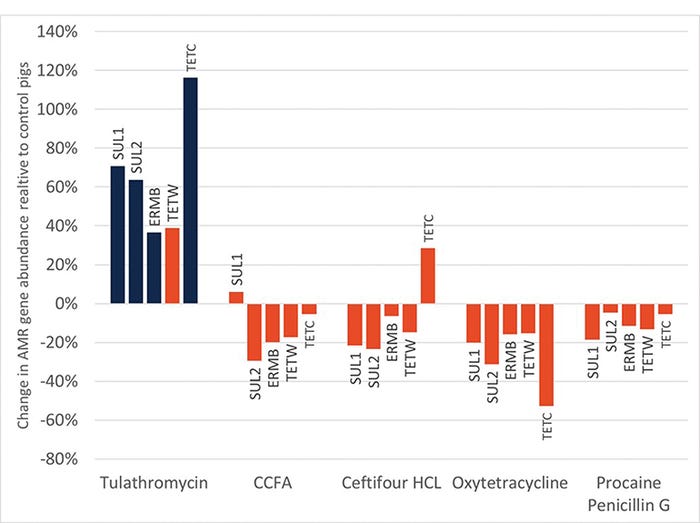
Figure 3: Relative abundance of antimicrobial resistance genes relative to control samples in piglets given various antibiotics at birth. Only one antibiotic had a significant impact (blue bars) on the relative abundance of ARG during the neonatal period.
While this is interesting, remember that antimicrobial resistance is a complex process, and increases in single genes may not reflect the entire picture of host health. To further investigate the TUL-treated pigs and determine the significance of the increased prevalence of the ARG, we subjected the TUL and control samples to whole genome sequencing on the Illumina MiSeq platform to amplify and determine all of the genetic elements responsible for antimicrobial resistance.
In this second, more robust (and more expensive, which is why there were limited samples analyzed) analysis pipeline, we determined that while the resistance to different antibiotics changed over time, TUL administration had only a minor, not significant, influence on the development of antibiotic resistance (see Figure 4).
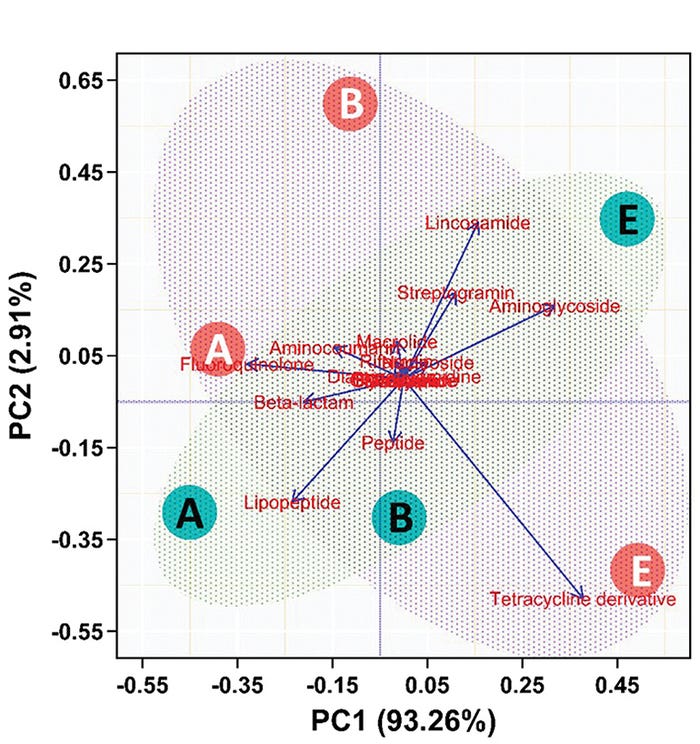
Figure 4: Principal component analysis of whole genome sequencing of swine fecal samples after administration in neonatal pigs. Resistance factors for classes of antibiotics were assessed for change over time (A=birth, B=5 days, C=20 days) and treatment (green=TUL, orange=control). The horizontal axis represents 93.3% of the total variation. The composition of the resistance factors changed with time (A, B and C are in same relative place on horizontal axis) but not treatment.
This more-robust analysis points out the fact that with these advanced, complicated approaches to analysis, how we conduct the analysis has a material impact on our findings. While we are not completely sure and further studies are needed, these data suggest that early antibiotic therapy in pigs has little impact on improved performance in the neonatal period or harm in producing more antimicrobial resistance. These results should be interpreted with caution, however, as these studies
were terminated at weaning, and there may be lifetime benefit or harm that could not be measured due to our approach.
How the microbiome develops over time has lasting impacts on the health of both humans and animals. If the development of the microbiome in people is shifted, it can result in altered digestive function, which can result in either malnutrition or obesity. These changes can be shaped by the diet (calorie restriction or high-calorie, low-quality diet),7 exposure to disease (diarrhea or excessive hygiene resulting in low bacterial exposure)8 or antibiotics.9
In cases of human malnutrition, a weeklong course of antibiotics has become the standard of care.10 The response to antibiotics in human malnutrition mimics the growth response seen in animals when we use oral antibiotics.11
There has been widespread focus on the role of antibiotics in the emergence of antibiotics resistance among bacteria in the host,11,12 due to medical use as well as use in farm animals and crops. Microbiome composition can be rapidly altered by exposure to antibiotics, with potential immediate effects on health — for instance, through the selection of resistant opportunistic pathogens that can cause acute disease.
Microbiome alterations induced by antibiotics can also indirectly affect long-term health. The mutualistic microbes in the human body interact with many physiological processes, and participate in the regulation of immune and metabolic homeostasis. Therefore, antibiotic exposure can alter many basic physiological equilibria, promoting long-term disease. In addition, excessive antibiotic use fosters bacterial resistance, and the overly exposed human microbiome has become a significant reservoir of resistance genes, contributing to the increasing difficulty in controlling bacterial infections.
Here, the complex relationships between antibiotics and the human microbiome are reviewed, with focus on the intestinal microbiota. Another important potential consequence of antimicrobial use is a negative impact on the structure and composition of the host microbiota.
It is important to understand that the gastrointestinal microbiota is not simply a transient population of microbes involved in nutrient metabolism, but that many microbial taxa coexist in a coordinated, complex mucosal ecosystem that contributes to host gastrointestinal and immunological development, particularly in the growing animal.13
If a healthy and stable microbiome is an important element of host health and development, then it is important to understand how common management practices, such as antimicrobial administration, might impact this complex host ecosystem in animals raised in intensive production systems.
There are multiple studies that investigate the use of oral antibiotics and their impact on the fecal microbiome in pigs. ASP 250 has been demonstrated to increase the relative abundance of E. coli in pig feces while increasing the expression of genes related to energy production and conversion.14 In another study, ASP 250 feeding in the immediate postweaning period resulted in no change in growth, but improved species richness and a lower prevalence of potential pathogens in the fecal material.15
There is a need of developing antibiotic growth promoter alternatives; however, the mechanisms by which AGP enhances livestock growth performance are not clearly understood. In this study, we fed 3-week-old swine for nine weeks with and without AGP containing chlortetracycline, sulfathiazole and penicillin to investigate the effects of AGPs on swine gut microbiota. Microbial community analysis was done based on bacterial 16S rRNA genes using MiSeq.
The use of AGP showed no growth-promoting effect, but inhibited the growth of potential pathogens during the early growth stage. Our results showed the significant increase in species richness after the stabilization of gut microbiota during the postweaning period (4-week-old).
When tylosin was fed to growing pigs over a 19-week period, there were significant shifts in the composition of the gut microbiome community that were not observed relative to controls in pigs fed chlortetracycline.16
Taken in total, these data suggest there are variable impacts on gut microbial community composition based on antibiotic class, but little is known about how these changes impact gut and host health.
Conversely, there is less known about the impact of injectable antibiotics on the swine gut microbiome. We conducted a study to characterize the impact of parenteral antibiotics administration on the composition and diversity of the resident fecal microbiota in growing pigs. Five antimicrobial treatment groups, each consisting of four 8-week-old piglets, were administered one of the antimicrobials — ceftiofur crystalline free acid, ceftiofur hydrochloride, oxytetracycline, procaine penicillin G and tulathromycin — at label dose and route. Individual fecal swabs were collected immediately before antimicrobial administration (control=Day 0), and again on days 1, 3, 7 and 14 after dosing. Genomic DNA was extracted, and the V1-V3 hypervariable region of 16S rRNA gene was amplified and sequenced using Illumina MiSeq-based sequencing.
Across all groups, the most abundant phyla were Firmicutes, Bacteroidetes and Proteobacteria. Linear discriminant analysis and stacked area graphs showed a pronounced, antimicrobial-dependent shift in the composition of fecal microbiota over time from Day 0.
By Day 14, the fecal microbial compositions of the groups receiving CHC and TUL had returned to a distribution that closely resembled that observed on Day 0, but differences were still evident.
In contrast, animals that received PPG, OTC and CCFA showed a tendency toward a balanced homeostatic microbiota structure on Day 7, but appeared to deviate away from the Day 0 composition by Day 14.
In conclusion, parenteral antimicrobials administration showed significant shifts in the composition and diversity of fecal microbiota in growing pigs. The observed changes in fecal microbiota structure showed antimicrobial-specific variations in both duration and extent. None of the groups exhibited a full return to preadministration fecal microbial community structure by Day 14 post-treatment.
In total, these data suggest there are likely both benefits and dangers for gut health when we give injectable antibiotics. As we gain a deeper understanding of what these effects are, we will be able to design treatment regimens focused on optimizing the health of the host by targeting both pathogens and optimizing the structure of the host microbiome.
In summary, antibiotics have powerful effects on the composition and structure of the pig’s gut microbiome. These changes are likely linked to changes in the health of both the intestine locally and the host systemically.
As we continue to apply advanced genomic techniques under controlled experimental conditions, we will gain a much deeper understanding of the intricate interplay between the host and its microbiome. This understanding will continue to shape how we think about optimizing host health as we shift from a pathogen-centric to a host-centric view of disease. This shift will be critical if we want to optimize the efficiency of animal-based food systems and feed 9 billion people by 2020.
References
1. Makino, H.; A. Kushiro; E. Ishikawa; et al. “Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota.” PLoS One. 2013;8(11). doi:10.1371/journal.pone.0078331
2. Dominguez-Bello, M.G.; E.K. Costello; M. Contreras; et al. “Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns.” Proc Natl Acad Sci. 2010;107(26):11971-11975. doi:10.1073/pnas.1002601107
3. Roduit, C.; S. Scholtens; J.C. De Jongste; et al. “Asthma at 8 years of age in children born by caesarean section.” Thorax. 2009;64(2):107-113. doi:10.1136/thx.2008.100875
4. Rautava, S.; R. Luoto; S. Salminen; E. Isolauri. “Microbial contact during pregnancy, intestinal colonization and human disease.” Nat Rev Gastroenterol Hepatol. 2012;9(10):565-576. doi:10.1038/nrgastro.2012.144
5. Bonifacio, E.; K. Warncke; C. Winkler; M. Wallner; A.G. Ziegler. “Cesarean section and interferon-induced helicase gene polymorphisms combine to increase childhood type 1 diabetes risk.” Diabetes. 2011;60(12):3300-3306. doi:10.2337/db11-0729
6. Verani, J.R.; S.J. Schrag. “Group B Streptococcal Disease in Infants: Progress in Prevention and Continued Challenges.” Clin Perinatol. 2010;37(2):375-392. doi:10.1016/J.CLP.2010.02.002
7. Turnbaugh, P.J.; R.E. Ley; M.A. Mahowald; V. Magrini; E.R. Mardis; J.I. Gordon. “An obesity-associated gut microbiome with increased capacity for energy harvest.” Nature. 2006;444(7122):1027-1131. doi:10.1038/nature05414
8. Sekirov, I.; N.M. Tam; M. Jogova; et al. “Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection.” Infect Immun. 2008;76(10):4726-4736. doi:10.1128/IAI.00319-08
9. Azad, M.B.; S.L. Bridgman; A.B. Becker; A.L. Kozyrskyj. “Infant antibiotic exposure and the development of childhood overweight and central adiposity.” Int J Obes. 2014;38(10):1290-1298. doi:10.1038/ijo.2014.119
10. Community-based Management of Severe Acute Malnutrition. “Nearly 20 million children under 5 suffer from severe acute malnutrition.” www.who.int/nutrition/topics/
Statement_community_based_man_sev_acute_mal_eng.pdf. Accessed March 21, 2018.
11. Allen, H.K.; T.B. Stanton. “Altered Egos: Antibiotic Effects on Food Animal Microbiomes.” Annu Rev Microbiol. 2014;68(1): 297-315. doi:10.1146/annurev-micro-091213-113052
12. Francino, M.P. “Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances.” Front Microbiol. 2016;6:1543. doi:10.3389/fmicb.2015.01543
13. Clemente, J.C.; L.K. Ursell; L.W. Parfrey; R Knight. “The Impact of the Gut Microbiota on Human Health: An Integrative View.” Cell. 2012;148(6):1258-1270. doi:10.1016/j.cell.2012.01.035
14. Looft, T.; T.A. Johnson; H.K. Allen; et al. “In-feed antibiotic effects on the swine intestinal microbiome.” Proc Natl Acad Sci U S A. 2012;109(5):1691-1696. doi:10.1073/pnas.1120238109
15. Unno, T.; J-M Kim; R.B. Guevarra; S.G. Nguyen. “Effects of antibiotic growth promoter and characterization of ecological succession in swine gut microbiota.” J Microbiol Biotechnol. 2015;25(4):431-438. www.ncbi.nlm.nih.gov/pubmed/25370726. Accessed March 21, 2018.
16. Holman, D.B.; M.R. Chénier. “Temporal changes and the effect of subtherapeutic concentrations of antibiotics in the gut microbiota of swine.” FEMS Microbiol Ecol. 2014;90(3):599-608. doi:10.1111/1574-6941.12419
You May Also Like



