Emerging Salmonella isolated from Midwest swine
Increase in the frequency of isolation of the Salmonella 4,[5],12:i:- serotype in humans may be paralleled by a similar increase in swine clinical samples received.
February 14, 2017

By Ehud Elnekave, Samuel Hong, Peter Davies and Julio Alvarez, University of Minnesota College of Veterinary Medicine Department of Veterinary Population Medicine; and Albert Rovira, University of Minnesota College of Veterinary Medicine Veterinary Diagnostic Laboratory
Non-typhoidal Salmonella enterica infections cause approximately one million human cases in the United States every year, with an average of 20,000 hospitalizations and 400 deaths. Most domesticated animal species can be sub-clinically infected with NTS and can therefore act as sources of infection.
S. enterica can also cause clinical disease in animals, particularly certain serotype-host species combinations such as S. Choleraesuis or S. Typhimurium in swine. Although many Salmonella infections in humans caused by NTS are self-limiting, in some cases antimicrobial treatment is needed to treat systemic spread of the bacteria, thus the increasing prevalence of antimicrobial resistance in NTS isolates of both human and animal origin in the recent years is of concern. Trends in serotypes and AMR in Salmonella recovered from different source (humans, animals and food) is monitored in the United States through the National Antimicrobial Resistance Monitoring System for Enteric Bacteria.
Since the late-’90s the prevalence of S. 4,[5],12:i:-, a variant of S. Typhimurium that lacks the second phase flagellar antigen, has increased in the United States and worldwide. In the United States, S. 4,[5],12:i:- has gone from rarely being reported to the fourth-most common serotype among human cases in 2013 (1), and has been associated with numerous foodborne outbreaks in the United States and Europe in the last 20 years. Swine is one of the potential reservoirs of NTS, and over the years multiple serotypes have been recovered at different frequencies from swine samples collected at farms and slaughterhouses. While considered the second most likely source of foodborne salmonellosis in Europe (2), the contribution of swine in the United States has been estimated to be much lower (2, 3). Still, swine is typically considered the reservoir most commonly associated with the S.4,[5],12:i:- serotype, since pork and pork products have been identified as the source of several outbreaks caused by this serotype. Nevertheless, S.4,[5],12:i:- has been rarely found in pig samples collected in the slaughterhouse as part of the NARMS program (eight out of 947 samples collected between 2006-11), so its true on-farm prevalence is not known (3).
In a study conducted recently in our lab (4), a subset of approximately 2,500 NTS isolates of swine origin recovered from clinical samples received at the Minnesota Veterinary Diagnostic Laboratory in 2006-15 was analyzed to assess the serotype and AMR distribution. This study confirmed that swine can be infected with a large variety of Salmonella serotypes. Serotypes Typhimurium, Derby, Agona and 4,[5],12:i:- accounted for 50% of the isolates investigated. We found that while the proportion of S. Typhimurium and S. Derby isolates decreased throughout the study period, the prevalence of S. Agona and S. 4,[5],12:i:- increased (Figure 1). Interestingly, those increasing serotypes (Agona and 4,[5],12:i:-) showed an increased proportion of isolates resistant to ceftiofur and enrofloxacin, compared with other serotypes (4). An increase in the proportion of multidrug resistant S. 4,[5],12:i:- has been also noted in isolates of human origin (5).
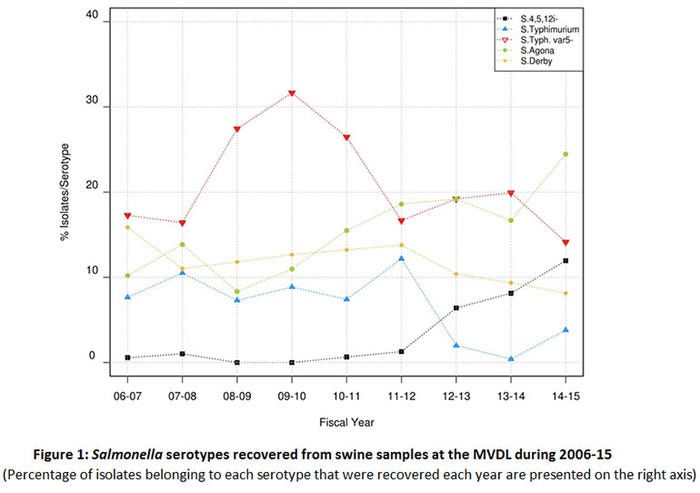
These results indicate that the increase in the frequency of isolation of the S.4,[5],12:i:- serotype in humans may be paralleled by a similar increase in swine clinical samples received in the MVDL. This is also supported by similar findings at the Iowa State University Diagnostic Laboratory (6). The rare isolation of S.4,[5],12:i:- in pig samples collected at the slaughterhouses indicates that risk for public health is potentially low, but further research is needed in order to identify the factors promoting the emergence of this serotype in humans and the role that swine may play as reservoirs.
References
1. Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual report; 2013.
2. Pires AF, Funk JA, Bolin C. Risk factors associated with persistence of Salmonella shedding in finishing pigs. Prev Vet Med. 2014 Sep 01;116(1-2):120-8.
3. Interagency Food Safety Analytics Collaboration Project. Foodborne Illness Source Attribution Estimates for Salmonella, Escherichia coli O157 (E. coli O157), Listeria monocytogenes (Lm), and Campylobacter using Outbreak Surveillance Data; 2015 Feburary.
4. Hong S, Rovira A, Davies P, Ahlstrom C, Muellner P, Rendahl A, et al. Serotypes and Antimicrobial Resistance in Salmonella enterica Recovered from Clinical Samples from Cattle and Swine in Minnesota, 2006 to 2015. PLoS One. 2016;11(12):e0168016.
5. National Antimicrobial Resistance Monitoring System. 2014 Human Isolates Surveillance Report: Centers for Disease Control and Prevention; 2014.
6. Krull A, Kreuder A, Arruda B, Burrough E, Schwartz K, Sahin O. Increased frequency of isolation of multi-drug resistant Salmonella I 4,[5],12:i:- from swine with histologic lesions consistent with salmonellosis. American Association of Veterinary Laboratory Diagnosticians Annual Conference; 2016; Greensborough, NC; 2016.
You May Also Like



